Viscoelastic differences between isolated and live MCF7 cancer cell nuclei resolved with AFM microrheology
- PMID: 40527475
- PMCID: PMC12173521
- DOI: 10.1098/rsif.2024.0885
Viscoelastic differences between isolated and live MCF7 cancer cell nuclei resolved with AFM microrheology
Abstract
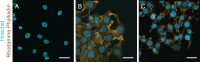
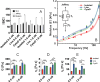
Cell nuclei are commonly isolated for mechanobiology studies although isolated nuclei may display viscoelastic properties differing from those of live cells. Nuclear mechanics is generally dependent on the time scale of the applied load and cannot accurately be assessed by a simple elasticity parameter. Active microrheology with an atomic force microscope (AFMMR) is a versatile tool for probing nuclear mechanics and we employ the technique for exploring isolated and live-cell nuclei in MCF7 cells, including the significance of actin depolymerization. We successfully validate the method using polyacrylamide hydrogels with correction for cantilever drag in the fluid. The AFMMR results reveal that isolated and live-cell nuclei are equivalent to within a scaling factor, in their frequency-dependent modulus, with isolated nuclei being softer. The loss tangent reveals a transition from solid- to liquid-like behaviour occurring at higher frequency in isolated than in live-cell nuclei. Viscoelastic modelling using the Jeffreys model describes the frequency-dependent modulus of all measured nuclei. Model parameters display sensitivity to nuclei isolation and to actin depolymerization in live cells. Sections of the Jeffreys circuit can potentially be assigned to internal and external nucleus structures, respectively, thereby establishing a minimal mechanistic framework for interpreting microrheology data on cell nuclei.
Keywords: MCF7; atomic force microscopy; cell nucleus; microrheology; viscoelasticity.
Conflict of interest statement
We declare we have no competing interests.
Figures





References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
