The Mycobacterium tuberculosis Transposon Sequencing Database (MtbTnDB): A Large-Scale Guide to Genetic Conditional Essentiality
- PMID: 40527579
- PMCID: PMC12242109
- DOI: 10.1111/mmi.15370
The Mycobacterium tuberculosis Transposon Sequencing Database (MtbTnDB): A Large-Scale Guide to Genetic Conditional Essentiality
Abstract
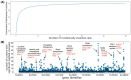
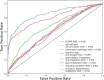
Characterizing genetic essentiality across various conditions is fundamental for understanding gene function. Transposon sequencing (TnSeq) is a powerful technique to generate genome-wide essentiality profiles in bacteria and has been extensively applied to Mycobacterium tuberculosis (Mtb). Dozens of TnSeq screens have yielded valuable insights into the biology of Mtb in vitro, inside macrophages, and in model host organisms. Despite their value, these Mtb TnSeq profiles have not been standardized or collated into a single, easily searchable database. This results in significant challenges when attempting to query and compare these resources, limiting our ability to obtain a comprehensive and consistent understanding of genetic conditional essentiality in Mtb. We address this problem by building a central repository of publicly available Mtb TnSeq screens, the Mtb transposon sequencing database (MtbTnDB). The MtbTnDB is a living resource that encompasses to date ≈150 standardized TnSeq screens, enabling open access to data, visualizations, and functional predictions through an interactive web app (www.mtbtndb.app). We conduct several statistical analyses on the complete database, such as demonstrating that (i) genes in the same genomic neighborhood have similar TnSeq profiles, and (ii) clusters of genes with similar TnSeq profiles are enriched for genes from similar functional categories. We further analyze the performance of machine learning models trained on TnSeq profiles to predict the functional annotation of orphan genes in Mtb. By facilitating the comparison of TnSeq screens across conditions, the MtbTnDB will accelerate the exploration of conditional genetic essentiality, provide insights into the functional organization of Mtb genes, and help predict gene function in this important human pathogen.
Keywords: database; functional genetics; microbiology; transposon sequencing; tuberculosis.
© 2025 The Author(s). Molecular Microbiology published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

