Circulating cell-free DNA methylation patterns indicate cellular sources of allograft injury after liver transplant
- PMID: 40527875
- PMCID: PMC12174327
- DOI: 10.1038/s41467-025-60507-9
Circulating cell-free DNA methylation patterns indicate cellular sources of allograft injury after liver transplant
Abstract
Post-transplant complications reduce allograft and recipient survival. Current approaches for detecting allograft injury non-invasively are limited and do not differentiate between cellular mechanisms. Here, we monitor cellular damages after liver transplants from cell-free DNA (cfDNA) fragments released from dying cells into the circulation. We analyzed 130 blood samples collected from 44 patients at different time points after transplant. Sequence-based methylation of cfDNA fragments were mapped to an atlas of cell-type-specific DNA methylation patterns derived from 476 methylomes of purified cells. For liver cell types, DNA methylation patterns and multi-omic data integration show distinct enrichment in open chromatin and functionally important regulatory regions. We find that multi-tissue cellular damages post-transplant recover in patients without allograft injury during the first post-operative week. However, sustained elevation of hepatocyte and biliary epithelial cfDNA within the first month indicates early-onset allograft injury. Further, cfDNA composition differentiates amongst causes of allograft injury indicating the potential for non-invasive monitoring and intervention.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: A.W., A.K., and M.E.M. are named as inventors on pending patent applications (U.S. Patent Application No. 18/291,113 and U.S. Patent Application No. 63/714,126) filed by Georgetown University, which cover the detection of liver damage using the methods described in this manuscript. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. Ethics approval and consent to participate: All collaborators of this study have fulfilled the criteria for authorship required by Nature Portfolio journals and have been included as authors, as their participation was essential for the design and implementation of the study. Roles and responsibilities were agreed among collaborators ahead of the research. This work includes findings that are locally relevant, which have been determined in collaboration with local partners. This research was not severely restricted or prohibited in the setting of the researchers, and does not result in stigmatization, incrimination, discrimination or personal risk to participants.
Figures


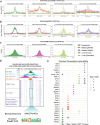

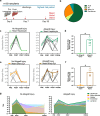

Update of
-
Circulating, cell-free methylated DNA indicates cellular sources of allograft injury after liver transplant.bioRxiv [Preprint]. 2024 Apr 5:2024.04.04.588176. doi: 10.1101/2024.04.04.588176. bioRxiv. 2024. Update in: Nat Commun. 2025 Jun 17;16(1):5310. doi: 10.1038/s41467-025-60507-9. PMID: 38617373 Free PMC article. Updated. Preprint.
References
-
- Lucey, M. R., Furuya, K. N. & Foley, D. P. Liver transplantation. N. Engl. J. Med.389, 1888–1900 (2023). - PubMed
-
- Craig, E. V. & Heller, M. T. Complications of liver transplant. Abdom. Radio.46, 43–67 (2021). - PubMed
-
- Limaye, A.R., Dixon, L.R. & Firpi, R.J. Liver biopsy - indications, procedures, results, 10.5772/52617 (2012).
-
- Heitzer, E., Auinger, L. & Speicher, M. R. Cell-free DNA and apoptosis: how dead cells inform about the living. Trends Mol. Med.26, 519–528 (2020). - PubMed
MeSH terms
Substances
Grants and funding
- T32-CA009686/U.S. Department of Health & Human Services | NIH | National Cancer Institute (NCI)
- P30-CA51008/U.S. Department of Health & Human Services | NIH | National Cancer Institute (NCI)
- R01 CA231291/CA/NCI NIH HHS/United States
- R01-AI132389/U.S. Department of Health & Human Services | NIH | National Institute of Allergy and Infectious Diseases (NIAID)
- P30 CA051008/CA/NCI NIH HHS/United States
- F30-CA250307/U.S. Department of Health & Human Services | NIH | National Cancer Institute (NCI)
- T32 CA009686/CA/NCI NIH HHS/United States
- R01-CA231291/U.S. Department of Health & Human Services | NIH | National Cancer Institute (NCI)
- F30 CA250307/CA/NCI NIH HHS/United States
- R01 AI132389/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical

