Nuclear deformability increases PARPi sensitivity in BRCA1-deficient cells by increasing microtubule-dependent DNA break mobility
- PMID: 40527886
- PMCID: PMC12174318
- DOI: 10.1038/s41467-025-60756-8
Nuclear deformability increases PARPi sensitivity in BRCA1-deficient cells by increasing microtubule-dependent DNA break mobility
Abstract
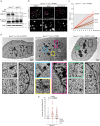
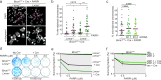
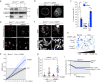
Microtubules and nuclear transmembrane SUN1/2 proteins promote the mobility of DNA Double Strand Breaks (DSBs) induced by ionizing radiation and the misrepair of one-ended DSBs induced in BRCA1-deficient cells by Poly(ADP-ribose) polymerase inhibitors (PARPi). However, whether microtubules promote aberrant DSBs repair by altering the nuclear structure and whether the nuclear structure itself plays a role in these processes is still unclear. Here we show that microtubule-dependent DSBs mobility in BRCA1-deficient cells after PARPi treatment is associated with nuclear envelope (NE) invaginations. Furthermore, increasing NE invaginations by Lmna deletion or inhibition of sphingolipid synthesis increases DSBs mobility, chromosomal aberrations, and PARPi cytotoxicity in BRCA1-deficient cells. These findings reveal a functional connection between the NE and DSB repair and suggest that drugs increasing NE deformability will enhance PARPi therapy efficacy in BRCA1-deficient cancers.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures


References
-
- Farmer, H. et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature434, 917–921 (2005). - PubMed
-
- Pommier, Y., O’Connor, M. J. & de Bono, J. Laying a trap to kill cancer cells: PARP inhibitors and their mechanisms of action. Sci. Transl. Med.8, 362ps17 (2016). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

