Skin biopsy processing for rapid molecular diagnosis and histopathologic interpretation: application to Kaposi sarcoma in East Africa
- PMID: 40528209
- PMCID: PMC12175338
- DOI: 10.1186/s13027-025-00671-1
Skin biopsy processing for rapid molecular diagnosis and histopathologic interpretation: application to Kaposi sarcoma in East Africa
Abstract
Background: Kaposi sarcoma (KS) is a cancer of viral origin (Kaposi sarcoma-associated herpesvirus; KSHV) for which the detection of KSHV DNA is an attractive target for a rapid, automatable diagnostic test. We previously demonstrated favorable diagnostic accuracy using loop-mediated isothermal amplification (LAMP) to quantitate KSHV DNA in lesional skin biopsies, though extracting DNA from the punch biopsies was the time-limiting step. Herein, we describe the development of a biopsy processing tool called Slicer to enable rapid nucleic acid testing in addition to traditional histopathological interpretation.
Methods: Slicer divides skin punch biopsies into two ½-cylinders and a thin, cross-sectional slice. The thin slice enables a previously demonstrated, equipment-free alkaline extraction termed ColdSHOT while the remaining ½-cylinders are available for histopathological diagnosis and additional molecular testing as needed. Slicer prototypes were used on skin punch biopsies collected from patients in Uganda who were referred for clinical suspicion of KS.
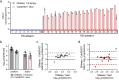
Results: For 27 patient samples, the combination of Slicer and ColdSHOT sample processing with LAMP testing resulted in qualitative KSHV DNA detection that was fully concordant with US-based histopathological diagnoses. Additional analysis demonstrated compatibility of Slicer and ColdSHOT with qPCR for KSHV DNA quantitation.
Conclusions: These results warrant further investigation using a larger set of skin biopsies and indicate that the Slicer and ColdSHOT could enable accurate KS diagnosis within a few hours of biopsy collection with minimal equipment.
Keywords: Kaposi sarcoma; Nucleic acid testing; Point-of-care testing; Skin sample processing.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: IRB approval was provided by Cornell University under IRB 0006836. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures




References
Grants and funding
LinkOut - more resources
Full Text Sources

