Cross-cohort analysis identifies shared gut microbial signatures and validates microbial risk scores for colorectal cancer
- PMID: 40528214
- PMCID: PMC12175378
- DOI: 10.1186/s12967-025-06676-z
Cross-cohort analysis identifies shared gut microbial signatures and validates microbial risk scores for colorectal cancer
Abstract
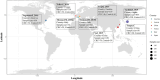
Background: Microbiome-wide association studies showed links between colorectal cancer (CRC) and gut microbiota. However, the clinical application of gut microbiota in CRC prevention has been hindered by the diversity of study populations and technical variations. We aimed to determine CRC-related gut microbial signatures based on cross-regional, cross-population, and cross-cohort metagenomic datasets, and elucidate its application value in CRC risk assessment.
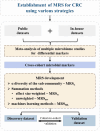
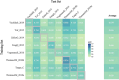
Methods: We used the MMUPHin tool to perform a meta-analysis of our own cohort and seven publicly available metagenomics datasets to identify gut microbial species associated with CRC across different cohorts, comprising of 570 CRC cases and 557 controls. Based on differential species sets, we constructed the microbial risk score (MRS) using α-diversity of the sub-community (MRSα), weighted/unweighted summation methods and machine learning algorithms. Cohort-to-cohort training and validation were performed to demonstrate the transferability.
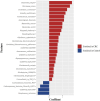
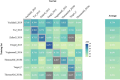
Results: We found that MRSα of core species was better validated and more interpretable than those constructed with summation methods or machine learning algorithms. Six species, including Parvimonas micra, Clostridium symbiosum, Peptostreptococcus stomatis, Bacteroides fragilis, Gemella morbillorum, and Fusobacterium nucleatum, were included in MRSα constructed by half or more of the cohorts. The AUC of MRSα, calculated based on the sub-community of six species, varied between 0.619 and 0.824 across the eight cohorts.
Conclusion: We identified six CRC-related species across regions, populations, and cohorts. The constructed MRSα could contribute to the risk prediction of CRC in different populations.
Keywords: Colorectal cancer; Cross-cohort; Disease prediction; Gut microbiota; Microbial risk score.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Data use from the Target-C study was approved by the Ethics Committee of the National Cancer Center/Cancer Hospital, Chinese Academy of Medical Sciences, and Peking Union Medical College (18-013/1615). All the participants provided written informed consent. Competing interests: The authors declare no conflict of interest.
Figures
Similar articles
-
The characteristics of tissue microbiota in different anatomical locations and different tissue types of the colorectum in patients with colorectal cancer.mSystems. 2025 Jun 17;10(6):e0019825. doi: 10.1128/msystems.00198-25. Epub 2025 May 27. mSystems. 2025. PMID: 40422085 Free PMC article.
-
Deciphering Gut Microbiome in Colorectal Cancer via Robust Learning Methods.Genes (Basel). 2025 Apr 15;16(4):452. doi: 10.3390/genes16040452. Genes (Basel). 2025. PMID: 40282413 Free PMC article.
-
Meta-analysis of gut microbiome reveals patterns of dysbiosis in colorectal cancer patients.J Med Microbiol. 2025 Jul;74(7):002042. doi: 10.1099/jmm.0.002042. J Med Microbiol. 2025. PMID: 40737178 Free PMC article.
-
Gut microbiota-derived metabolites in CRC progression and causation.J Cancer Res Clin Oncol. 2021 Nov;147(11):3141-3155. doi: 10.1007/s00432-021-03729-w. Epub 2021 Jul 17. J Cancer Res Clin Oncol. 2021. PMID: 34273006 Free PMC article. Review.
-
Bacteria-tumor symbiosis destructible novel nanocatalysis drug delivery systems for effective tumor therapy.Nanomedicine (Lond). 2025 Feb;20(3):305-318. doi: 10.1080/17435889.2024.2443388. Epub 2024 Dec 30. Nanomedicine (Lond). 2025. PMID: 39889806 Review.
References
-
- Bray F, Laversanne M, Sung H, Ferlay J, Siegel RL, Soerjomataram I, et al. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2024;74:229–63. - PubMed
-
- Siegel RL, Wagle NS, Cercek A, Smith RA, Jemal A. Colorectal cancer statistics, 2023. CA Cancer J Clin. 2023;73:233–54. - PubMed
-
- Ladabaum U, Dominitz JA, Kahi C, Schoen RE. Strategies for colorectal cancer screening. Gastroenterology. 2020;158:418–32. - PubMed
-
- Kaminski MF, Robertson DJ, Senore C, Rex DK. Optimizing the quality of colorectal cancer screening worldwide. Gastroenterology. 2020;158:404–17. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

