Structural variation detection and association analysis of whole-genome-sequence data from 16,543 Alzheimer's disease sequencing project subjects
- PMID: 40528303
- PMCID: PMC12173831
- DOI: 10.1002/alz.70277
Structural variation detection and association analysis of whole-genome-sequence data from 16,543 Alzheimer's disease sequencing project subjects
Abstract
Introduction: The role of structural variations (SVs) in Alzheimer's disease (AD) remains understudied.
Methods: We analyzed whole-genome sequencing data from the Alzheimer's Disease Sequencing Project (N = 16,543) and identified 400,234 (168,223 high-quality) SVs. Laboratory validation yielded a sensitivity of 82% (85% for high-quality).
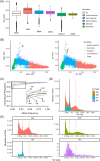
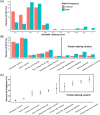
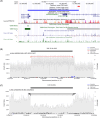
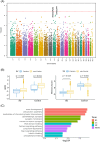
Results: We found a burden of singletons (odds ratio [OR] = 1.07, p = 0.0017) and homozygous deletions (OR = 1.14, p < 0.0001) in cases. On AD genes, we observed the ultra-rare SVs associated with the disease, including protein-altering SVs in ABCA7, APP, PLCG2, and SORL1. Twenty-one SVs are in linkage disequilibrium (LD) with known AD-risk variants, exemplified by a 5k deletion in LD (R2 = 0.99) with rs143080277 in NCK2. We identified a rare deletion near RNA5SP293 associated with AD (OR = 1.99, p = 1.3 × 10-5), which was replicated using an independent dataset.
Discussion: This study highlights the pivotal role of SVs in AD genetics.
Highlights: Observed a significant burden of singletons and homozygous deletions in Alzheimer's disease (AD) patients. Identified rare protein-altering structural variations (SVs) in ABCA7, APP, PLCG2, and SORL1. Established linkages between SVs and AD risk-associated single nucleotide variants (SNVs). Discovered a novel deletion near RNA5SP293 linked to AD, replicated independently. Uncovered over-representation of SVs in neuronal function pathways.
Keywords: Alzheimer's disease; NCK2; RNA5SP293; copy number variation; structural variation.
© 2025 The Author(s). Alzheimer's & Dementia published by Wiley Periodicals LLC on behalf of Alzheimer's Association.
Conflict of interest statement
The authors declare no conflicts of interest. Author disclosures are available in Supporting Information.
Figures
Update of
-
Structural Variation Detection and Association Analysis of Whole-Genome-Sequence Data from 16,905 Alzheimer's Diseases Sequencing Project Subjects.medRxiv [Preprint]. 2023 Sep 13:2023.09.13.23295505. doi: 10.1101/2023.09.13.23295505. medRxiv. 2023. Update in: Alzheimers Dement. 2025 Jun;21(6):e70277. doi: 10.1002/alz.70277. PMID: 37745545 Free PMC article. Updated. Preprint.
-
Structural Variation Detection and Association Analysis of Whole-Genome-Sequence Data from 16,905 Alzheimer's Diseases Sequencing Project Subjects.Res Sq [Preprint]. 2023 Oct 5:rs.3.rs-3353179. doi: 10.21203/rs.3.rs-3353179/v1. Res Sq. 2023. Update in: Alzheimers Dement. 2025 Jun;21(6):e70277. doi: 10.1002/alz.70277. PMID: 37886469 Free PMC article. Updated. Preprint.
Similar articles
-
Structural Variation Detection and Association Analysis of Whole-Genome-Sequence Data from 16,905 Alzheimer's Diseases Sequencing Project Subjects.Res Sq [Preprint]. 2023 Oct 5:rs.3.rs-3353179. doi: 10.21203/rs.3.rs-3353179/v1. Res Sq. 2023. Update in: Alzheimers Dement. 2025 Jun;21(6):e70277. doi: 10.1002/alz.70277. PMID: 37886469 Free PMC article. Updated. Preprint.
-
Structural Variation Detection and Association Analysis of Whole-Genome-Sequence Data from 16,905 Alzheimer's Diseases Sequencing Project Subjects.medRxiv [Preprint]. 2023 Sep 13:2023.09.13.23295505. doi: 10.1101/2023.09.13.23295505. medRxiv. 2023. Update in: Alzheimers Dement. 2025 Jun;21(6):e70277. doi: 10.1002/alz.70277. PMID: 37745545 Free PMC article. Updated. Preprint.
-
Structural variants linked to Alzheimer's disease and other common age-related clinical and neuropathologic traits.Genome Med. 2025 Mar 4;17(1):20. doi: 10.1186/s13073-025-01444-6. Genome Med. 2025. PMID: 40038788 Free PMC article.
-
18F PET with florbetapir for the early diagnosis of Alzheimer's disease dementia and other dementias in people with mild cognitive impairment (MCI).Cochrane Database Syst Rev. 2017 Nov 22;11(11):CD012216. doi: 10.1002/14651858.CD012216.pub2. Cochrane Database Syst Rev. 2017. PMID: 29164603 Free PMC article.
-
Vitamin E for Alzheimer's dementia and mild cognitive impairment.Cochrane Database Syst Rev. 2017 Apr 18;4(4):CD002854. doi: 10.1002/14651858.CD002854.pub5. Cochrane Database Syst Rev. 2017. PMID: 28418065 Free PMC article.
Cited by
-
The role of peripheral innate immune cells in Alzheimer's disease progression.Front Immunol. 2025 Jul 16;16:1616939. doi: 10.3389/fimmu.2025.1616939. eCollection 2025. Front Immunol. 2025. PMID: 40740772 Free PMC article. Review.
-
A Specialized Reference Panel with Structural Variants Integration for Improving Genotype Imputation in Alzheimer's Disease and Related Dementias (ADRD).medRxiv [Preprint]. 2024 Jul 23:2024.07.22.24310827. doi: 10.1101/2024.07.22.24310827. medRxiv. 2024. Update in: HGG Adv. 2025 Jul 31;6(4):100487. doi: 10.1016/j.xhgg.2025.100487. PMID: 39108532 Free PMC article. Updated. Preprint.
-
Alzheimer's Disease Sequencing Project Release 4 Whole Genome Sequencing Dataset.medRxiv [Preprint]. 2024 Dec 6:2024.12.03.24317000. doi: 10.1101/2024.12.03.24317000. medRxiv. 2024. Update in: Alzheimers Dement. 2025 May;21(5):e70237. doi: 10.1002/alz.70237. PMID: 39677464 Free PMC article. Updated. Preprint.
References
-
- Gaugler J, James B, Johnson T, et al. 2022 Alzheimer's disease facts and figures. Alzheimers Dement. 2022;18(4):700‐789. - PubMed
-
- Martins RN, Clarnette R, Fisher C, et al. ApoE genotypes in Australia: roles in early and late onset Alzheimer's disease and Down's syndrome. Neuroreport. 1995;6(11):1513‐1516. - PubMed
MeSH terms
Substances
Grants and funding
- RF1 AG074328/AG/NIA NIH HHS/United States
- U01-AG032984/AG/NIA NIH HHS/United States
- U24 AG041689/AG/NIA NIH HHS/United States
- R01-AG048927/AG/NIA NIH HHS/United States
- R01 AG074328/AG/NIA NIH HHS/United States
- U01 AG062602/AG/NIA NIH HHS/United States
- R01 AG048927/AG/NIA NIH HHS/United States
- U54-AG052427/AG/NIA NIH HHS/United States
- P30-AG072978/AG/NIA NIH HHS/United States
- P30 AG072979/AG/NIA NIH HHS/United States
- P30AG072979/AG/NIA NIH HHS/United States
- U01 AG058654/AG/NIA NIH HHS/United States
- U01-AG062602/AG/NIA NIH HHS/United States
- U01-AG058654/AG/NIA NIH HHS/United States
- U54 AG052427/AG/NIA NIH HHS/United States
- R01-AG074328/AG/NIA NIH HHS/United States
- U01 AG032984/AG/NIA NIH HHS/United States
- P30 AG072978/AG/NIA NIH HHS/United States
- U24-AG041689/AG/NIA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Research Materials

