Single-Cell Lipidomics by LC-MS Interlaboratory Study Reveals the Impact of X-ray Irradiation on a Pancreatic Cancer Cell Line and Its Bystanders
- PMID: 40528311
- PMCID: PMC12224164
- DOI: 10.1021/acs.analchem.5c02010
Single-Cell Lipidomics by LC-MS Interlaboratory Study Reveals the Impact of X-ray Irradiation on a Pancreatic Cancer Cell Line and Its Bystanders
Abstract
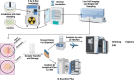
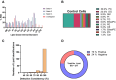
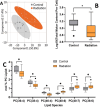
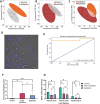
Live single-cell lipidomics by liquid chromatography mass spectrometry (LC-MS) is a nascent and rapidly growing field which can shed new light on infectious diseases, cancer, immunology, and drug delivery. There are now a growing number of laboratories that can isolate single cells and laboratories that can perform lipidomics analysis at correspondingly low sample volumes, but there is a lack of validation data. We have carried out the first interlaboratory LC-MS lipidomics experiment for single cells, aimed at filling this gap. We present a novel workflow to enable interlaboratory studies, comprising live-cell imaging and single-cell isolation, followed by freeze-drying, international shipping, reconstitution, and untargeted lipidomics analysis. We applied this methodology to reveal radiation-induced bystander effects in pancreatic cancer cells. X-ray irradiated cells and their bystanders sampled live 48 h postirradiation demonstrated reduced lipid abundance compared to controls, with distinct changes in molar ratios of several polyunsaturated lipids. This demonstrates for the first time that radiation can cause considerable cellular lipid remodelling, not only at the site of delivery. A striking similarity in lipid changes was observed between the two participating laboratories despite differences in sample preparation and analysis methods. Our results are further corroborated by live-cell imaging analysis of lipid droplets. This work serves as an important validation and demonstration of the nascent and rapidly growing field of live single-cell lipidomics.
Figures

Similar articles
-
A rapid and systematic review of the clinical effectiveness and cost-effectiveness of paclitaxel, docetaxel, gemcitabine and vinorelbine in non-small-cell lung cancer.Health Technol Assess. 2001;5(32):1-195. doi: 10.3310/hta5320. Health Technol Assess. 2001. PMID: 12065068
-
Signs and symptoms to determine if a patient presenting in primary care or hospital outpatient settings has COVID-19.Cochrane Database Syst Rev. 2022 May 20;5(5):CD013665. doi: 10.1002/14651858.CD013665.pub3. Cochrane Database Syst Rev. 2022. PMID: 35593186 Free PMC article.
-
Electronic cigarettes for smoking cessation.Cochrane Database Syst Rev. 2021 Sep 14;9(9):CD010216. doi: 10.1002/14651858.CD010216.pub6. Cochrane Database Syst Rev. 2021. Update in: Cochrane Database Syst Rev. 2022 Nov 17;11:CD010216. doi: 10.1002/14651858.CD010216.pub7. PMID: 34519354 Free PMC article. Updated.
-
Interventions to prevent occupational noise-induced hearing loss.Cochrane Database Syst Rev. 2017 Jul 7;7(7):CD006396. doi: 10.1002/14651858.CD006396.pub4. Cochrane Database Syst Rev. 2017. PMID: 28685503 Free PMC article.
-
Electronic cigarettes for smoking cessation.Cochrane Database Syst Rev. 2022 Nov 17;11(11):CD010216. doi: 10.1002/14651858.CD010216.pub7. Cochrane Database Syst Rev. 2022. Update in: Cochrane Database Syst Rev. 2024 Jan 8;1:CD010216. doi: 10.1002/14651858.CD010216.pub8. PMID: 36384212 Free PMC article. Updated.
References
-
- Wang Z., Cao M., Lam S. M., Shui G.. Embracing Lipidomics at Single-Cell Resolution: Promises and Pitfalls. TrAC Trends in Analytical Chemistry. 2023;160:116973. doi: 10.1016/j.trac.2023.116973. - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

