Identification of technically challenging variants: Whole-genome sequencing improves diagnostic yield in patients with high clinical suspicion of rare diseases
- PMID: 40528347
- PMCID: PMC12269974
- DOI: 10.1016/j.xhgg.2025.100469
Identification of technically challenging variants: Whole-genome sequencing improves diagnostic yield in patients with high clinical suspicion of rare diseases
Abstract
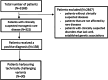
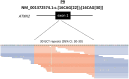
The total burden of rare diseases is significant worldwide, with over 300 million people being affected. Many rare diseases have both well-defined clinical phenotypes and established genetic causes. However, a remarkable proportion of patients with high clinical suspicion of a rare disease remain genetically undiagnosed and stuck in the diagnostic odyssey after having a cascade of conventional genetic tests. One of the major factors contributing to this is that many types of variants are technically intractable to whole-exome sequencing (WES). In this study, the added diagnostic power of whole-genome sequencing (WGS) for patients with clinically suspected rare diseases was assessed by detecting technically challenging variants. 3,169 patients from the Hong Kong Genome Project (HKGP) were reviewed, identifying 322 individuals having high clinical suspicion of a rare disorder with well-established genetic etiology. Notably, 180 patients have performed at least one previous genetic test. Through PCR-free short-read WGS and a comprehensive in-house analytic pipeline, causative variants were found in 138 patients (138 of 322, 42.9%), 30 of which (30 of 138, 21.7%) are attributed to technically challenging variants. These included 6 variants in low-coverage regions with PCR bias, 2 deep intronic variants, 2 repeat expansions, 19 structural variants, and 2 variants in genes with a homologous pseudogene. The study demonstrated the indispensable diagnostic power of WGS in detecting technically challenging variants and the capability to serve as an all-in-one test for patients with high clinical suspicion of rare diseases.
Keywords: challenging variants; non-coding variants; pseudogenes; rare diseases; repeat expansions; structural variants; whole genome sequencing.
Copyright © 2025 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests W.-K.J.L. is a director of DRA.
Figures




References
-
- Lincoln S.E., Hambuch T., Zook J.M., Bristow S.L., Hatchell K., Truty R., Kennemer M., Shirts B.H., Fellowes A., Chowdhury S., et al. One in seven pathogenic variants can be challenging to detect by NGS: an analysis of 450,000 patients with implications for clinical sensitivity and genetic test implementation. Genet. Med. 2021;23:1673–1680. doi: 10.1038/s41436-021-01187-w. - DOI - PMC - PubMed
-
- AlAbdi L., Maddirevula S., Shamseldin H.E., Khouj E., Helaby R., Hamid H., Almulhim A., Hashem M.O., Abdulwahab F., Abouyousef O., et al. Diagnostic implications of pitfalls in causal variant identification based on 4577 molecularly characterized families. Nat. Commun. 2023;14:5269. doi: 10.1038/s41467-023-40909-3. - DOI - PMC - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

