The genome sequence of the Asparagus Beetle, Crioceris asparagi (Linnaeus, 1758)
- PMID: 40528997
- PMCID: PMC12171721
- DOI: 10.12688/wellcomeopenres.23460.1
The genome sequence of the Asparagus Beetle, Crioceris asparagi (Linnaeus, 1758)
Abstract
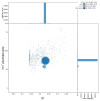
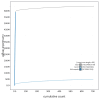
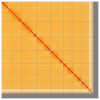
We present a genome assembly from an individual male specimen of Crioceris asparagi (Asparagus Beetle; Arthropoda; Insecta; Coleoptera; Chrysomelidae). The genome sequence has a total length of 639.30 megabases. Most of the assembly (93.04%) is scaffolded into 9 chromosomal pseudomolecules, including the X and Y sex chromosomes. The mitochondrial genome has also been assembled and is 15.76 kilobases in length. Gene annotation of this assembly on Ensembl identified 26,673 protein-coding genes.
Keywords: Asparagus Beetle; Coleoptera; Crioceris asparagi; chromosomal; genome sequence.
Copyright: © 2025 Geiser MF et al.
Conflict of interest statement
No competing interests were disclosed.
Figures


Similar articles
-
The genome sequence of a flea beetle, Altica lythri Aubé, 1843.Wellcome Open Res. 2025 Jun 2;10:297. doi: 10.12688/wellcomeopenres.24269.1. eCollection 2025. Wellcome Open Res. 2025. PMID: 40931741 Free PMC article.
-
The genome sequence of a soldier beetle, Cantharis flavilabris Fallén, 1807.Wellcome Open Res. 2024 Oct 15;9:303. doi: 10.12688/wellcomeopenres.22422.2. eCollection 2024. Wellcome Open Res. 2024. PMID: 39280726 Free PMC article.
-
The genome sequence of a ground beetle, Harpalus rufipes (DeGeer, 1774).Wellcome Open Res. 2024 Aug 14;9:483. doi: 10.12688/wellcomeopenres.22875.1. eCollection 2024. Wellcome Open Res. 2024. PMID: 40861021 Free PMC article.
-
The genome sequence of a soldier fly, Nemotelus pantherinus (Linnaeus, 1758).Wellcome Open Res. 2025 Jun 2;10:303. doi: 10.12688/wellcomeopenres.24309.1. eCollection 2025. Wellcome Open Res. 2025. PMID: 40548330 Free PMC article.
-
The genome sequence of a phantom cranefly, Ptychoptera contaminata (Linnaeus, 1758).Wellcome Open Res. 2025 Jun 4;10:308. doi: 10.12688/wellcomeopenres.24276.1. eCollection 2025. Wellcome Open Res. 2025. PMID: 40823656 Free PMC article.
References
-
- Bates A, Clayton-Lucey I, Howard C: Sanger Tree of Life HMW DNA fragmentation: diagenode Megaruptor ®3 for LI PacBio. protocols.io. 2023. 10.17504/protocols.io.81wgbxzq3lpk/v1 - DOI
-
- Beasley J, Uhl R, Forrest LL, et al. : DNA barcoding SOPs for the Darwin Tree of Life project. protocols.io. 2023; [Accessed 25 June 2024]. 10.17504/protocols.io.261ged91jv47/v1 - DOI
-
- Bezděk J, Sekerka L: Catalogue of palaearctic coleoptera. Volume 6/2/1. Updated and revised second edition. Chrysomeloidea II (Orsodacnidae, Megalopodidae, Chrysomelidae).Leiden: Brill,2024. 10.1163/9789004443303 - DOI
Grants and funding
LinkOut - more resources
Full Text Sources

