Integrated pan-cancer analysis of ADM's role in prognosis, immune modulation and resistance
- PMID: 40529377
- PMCID: PMC12170686
- DOI: 10.3389/fimmu.2025.1573250
Integrated pan-cancer analysis of ADM's role in prognosis, immune modulation and resistance
Abstract
Introduction: Adrenomedullin (ADM), a multifunctional peptide, has been implicated in various inflammatory and autoimmune diseases. However, its role in cancer, particularly in NSCLC, remained under-explored. This called for a pan-cancer analysis of ADM, investigating its expression, genomic alterations, prognostic value, immune associations, and relations with drug sensitivity to provide insights into its potential as a therapeutic target and biomarker.
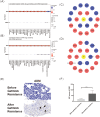
Methods: ADM expression data from normal and tumor tissues was retrieved and analyzed through HPA and Timer 2.0 online platforms. Genetic alterations, copy number variations (CNVs), and methylation patterns were analyzed using cBioPortal and GSCA platforms. The data for survival analysis was extracted from TCGA and GEO database and analyzed through GEPIA and PrognoScan online platforms. ADM's correlations with immune checkpoint genes, immune cell infiltration, MSI, and TMB were evaluated using data from Timer and TCGA via R. Drug sensitivity analysis was performed with GDSC and CTRP databases, supported by network visualizations. IHC staining was conducted on LUAD patients' samples to assess ADM's relationship with EGFR-TKI resistance and immune microenvironment.
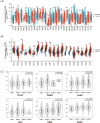
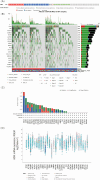
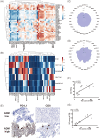
Results: ADM was widely expressed across normal tissues, with high levels in adipose tissue, endocrine organs, digestive and reproductive systems. Pan-cancer analysis revealed that ADM expression was upregulated in multiple cancer types, including CESC, ESCA, GBM, HNSC, KICH, KIRC, LUSC, PCPG, THCA, and UCEC, and correlated with advanced pathological stages in THCA, KIRP, and HNSC. Furthermore, high ADM expression was significantly linked to poor prognosis in patients with LGG, LUAD, MESO, THYM, LIHC, HNSC, GBM, KICH, KIRP, CESC, PAAD, and STAD, while its negative influence on OS and RFS was validated in LUAD. In addition, ADM exhibited genetic alterations, including amplification and deep deletion across multiple cancer types. Strong and consistent positive correlations were witnessed between ADM and several immune checkpoint genes, including CD274 (PD-L1), CD276, TNFRSF18, TNFSF9, and PVR in pan-cancer analysis, indicating its role in the development of suppressive immune microenvironment and T cell exhaustion. Besides, ADM showed significant correlations with immune cell infiltration, and TMB/MSI, highlighting its role in immune regulation and its potential as a predictive biomarker for immunotherapy. Significantly, ADM expression was correlated with multiple drug sensitivity, particularly chemotherapy and tyrosine kinase inhibitors (TKIs) therapy. Moreover, positive correlations between its expression and EGFR-TKI resistance, CD8+ T cell infiltration and tumor proportion score (TPS) in LUAD were validated in patients' samples, emphasizing its potential in guiding personalized therapy.
Discussion: This pan-cancer analysis revealed ADM's pivotal role in progression, immune modulation, and therapeutic response, especially in LUAD. ADM held promise as a prognostic biomarker and a potential therapeutic target in immune modulation and resistance management. Future research should focus on experimental validation and elucidation of ADM-mediated pathways, which might provide novel insights into cancer biology and improve clinical outcomes.
Keywords: adrenomedullin (ADM); immune modulation; lung adenocarcinoma (LUAD); pan-cancer analysis; resistance.
Copyright © 2025 Liu and Zhou.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




Similar articles
-
Interplay between tumor mutation burden and the tumor microenvironment predicts the prognosis of pan-cancer anti-PD-1/PD-L1 therapy.Front Immunol. 2025 Jul 24;16:1557461. doi: 10.3389/fimmu.2025.1557461. eCollection 2025. Front Immunol. 2025. PMID: 40777041 Free PMC article.
-
Unraveling the role of GPCR signaling in metabolic reprogramming and immune microenvironment of lung adenocarcinoma: a multi-omics study with experimental validation.Front Immunol. 2025 Jun 6;16:1606125. doi: 10.3389/fimmu.2025.1606125. eCollection 2025. Front Immunol. 2025. PMID: 40547013 Free PMC article.
-
Comprehensive analysis of 33 human cancers reveals clinical implications and immunotherapeutic value of the solute carrier family 35 member A2.Front Immunol. 2023 May 18;14:1155182. doi: 10.3389/fimmu.2023.1155182. eCollection 2023. Front Immunol. 2023. PMID: 37275857 Free PMC article.
-
Pan-cancer and multiomics: advanced strategies for diagnosis, prognosis, and therapy in the complex genetic and molecular universe of cancer.Clin Transl Oncol. 2025 Jul;27(7):2936-2954. doi: 10.1007/s12094-024-03819-4. Epub 2024 Dec 26. Clin Transl Oncol. 2025. PMID: 39725831 Review.
-
Mechanisms, combination therapy, and biomarkers in cancer immunotherapy resistance.Cell Commun Signal. 2024 Jun 19;22(1):338. doi: 10.1186/s12964-024-01711-w. Cell Commun Signal. 2024. PMID: 38898505 Free PMC article. Review.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

