Overlapping and specific bacterial communities in the gut and reproductive system of Bactrocera dorsalis (Hendel) (Diptera: Tephritidae) adults
- PMID: 40529586
- PMCID: PMC12171230
- DOI: 10.3389/fmicb.2025.1567154
Overlapping and specific bacterial communities in the gut and reproductive system of Bactrocera dorsalis (Hendel) (Diptera: Tephritidae) adults
Abstract
Background: Different insect tissues represent heterogeneous niches with distinct physiological and biochemical characteristics, and therefore host different bacterial communities.
Methods: In this study, those overlapping and specific bacterial communities in the female gut (fG), male gut (mG), female reproductive system (fR), and male reproductive system (mR) of Bactrocera dorsalis (Hendel) adults were determined by high-throughput sequencing targeting 16S rRNA gene.
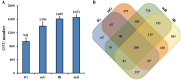
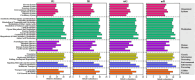
Results: The richness of bacterial taxa based on OTU was higher in fR compared to the other three tissues. Among the 29 identified bacterial phyla, Pseudomonadota, Bacillota, and Bacteroidota were predominant, while among the 48 identified genera, Enterobacter, Kluyvera, Asticcacaulis, Mesorhizobium, and Serratia were common in the four tissues. fG harbored specific bacterial genus Morganella, mG harbored specific bacterial genera Vagococcus, Lactobacillus, Lactococcus, Lactobacillales, and Bacilli, fR harbored specific bacterial genera Blastomonas, Ralstonia and Providencia, and mR harbored specific bacterial genera Sphingobacteriia, Asticcacaulis, Caulobacter, Caulobacterales, Bradyrhizobium, and Luteimonas. In the 35 annotated KEGG pathways, high-abundance bacterial taxa were mainly enriched in these pathways of membrane transport, carbohydrate metabolism, amino acid metabolism, replication and repair, and energy metabolism, while low-abundance bacterial taxa were involved in these pathways of cardiovascular diseases, circulatory system, and excretory system. The abundances of the 5 pathways associated with cardiovascular diseases, circulatory system, excretory system, membrane transport, and polysaccharide biosynthesis and metabolism exhibited greater variations among fG, mG, fR, and mR. Among them, the two pathways abundances of cardiovascular disease and circulatory system were higher in the reproductive system, whereas the other three pathways abundances were higher in the female gut.
Conclusion: Our study revealed the abundance, composition and function of overlapping and specific bacterial communities in the gut and reproductive system of B. dorsalis, providing valuable information for inhibiting the occurrence of B. dorsalis by interfering with these functional bacterial communities in tissues.
Keywords: Bactrocera dorsalis; bacterial diversity; function annotation; gut; reproductive system; specific bacterial communities.
Copyright © 2025 Li, Hu, Zhong, Song, Xu, Xie and Li.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




Similar articles
-
Bacterial communities in the gut of wild and mass-reared Zeugodacus cucurbitae and Bactrocera dorsalis revealed by metagenomic sequencing.BMC Microbiol. 2019 Dec 24;19(Suppl 1):282. doi: 10.1186/s12866-019-1647-8. BMC Microbiol. 2019. PMID: 31870295 Free PMC article.
-
Electroantennographic and behavioral responses of Bactrocera dorsalis (Diptera: Tephritidae) adults to the volatiles of plum fruits.J Econ Entomol. 2024 Dec 28;117(6):2400-2412. doi: 10.1093/jee/toae225. J Econ Entomol. 2024. PMID: 39393005
-
Integrating Gut Microbiome and Metabolomics with Magnetic Resonance Enterography to Advance Bowel Damage Prediction in Crohn's Disease.J Inflamm Res. 2025 Jun 11;18:7631-7649. doi: 10.2147/JIR.S524671. eCollection 2025. J Inflamm Res. 2025. PMID: 40535353 Free PMC article.
-
Metagenomics-assembled genomes reveal microbial metabolic adaptation to athalassohaline environment, the case Lake Barkol, China.Front Microbiol. 2025 Jun 4;16:1550346. doi: 10.3389/fmicb.2025.1550346. eCollection 2025. Front Microbiol. 2025. PMID: 40535002 Free PMC article.
-
A meta-analysis of genome-wide association studies to identify candidate genes associated with feed efficiency traits in pigs.J Anim Sci. 2025 Jan 4;103:skaf010. doi: 10.1093/jas/skaf010. J Anim Sci. 2025. PMID: 39847436 Free PMC article.
References
LinkOut - more resources
Full Text Sources

