Precision targeting of genetic variations in mixed bacterial cultures using CRISPR-Cas12a-programmed λ phages
- PMID: 40529587
- PMCID: PMC12171280
- DOI: 10.3389/fmicb.2025.1575339
Precision targeting of genetic variations in mixed bacterial cultures using CRISPR-Cas12a-programmed λ phages
Abstract
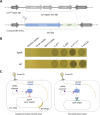
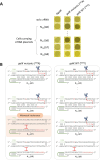
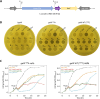
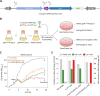
The CRISPR-Cas system, an adaptive immune mechanism in prokaryotes against bacteriophages, has been developed into a versatile tool for recognizing and cleaving target nucleic acid sequences. In this study, we developed a model system by integrating CRISPR-Cas12a into the genome of temperate bacteriophage λ, enabling precise regulation of lysogeny and lysis in Escherichia coli. We confirmed that λ phage, armed with Cas12a nuclease and CRISPR RNA (crRNA) targeting specific sequences, could inhibit the lysogenic cycle of E. coli cells. We demonstrated that the CRISPR-Cas12a-loaded temperate λ phage mimicked a lytic phage by selectively killing cells carrying the target genomic sequence. Furthermore, by employing truncated crRNA to enhance target recognition specificity, we found that the synthetic phage could distinguish single nucleotide variations in the genomic target DNA, enabling precise targeting and selective elimination of target cells in homogeneous bacterial cultures. To further validate its specificity, we tested this system in mixed bacterial cultures, wherein Cas12a nuclease and truncated crRNA-loaded bacteriophages selectively eliminated only those cells carrying the target sequences perfectly matching the crRNA. These results highlight the potential of this approach for advancing precision microbiome modulation.
Keywords: CRISPR-Cas12a; Escherichia coli; bacteriophage; lambda; lysogen.
Copyright © 2025 Lee, Lee, Jeong and Lee.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Figures
References
-
- Balcha F. B., Neja S. A. (2023). CRISPR-Cas9 mediated phage therapy as an alternative to antibiotics. Anim. Dis. 3, 1–12. doi: 10.1186/s44149-023-00065-z - DOI
LinkOut - more resources
Full Text Sources

