The mechanism of RNA methylation writing protein-related prognostic genes in lung adenocarcinoma based on bioinformatics
- PMID: 40529813
- PMCID: PMC12171300
- DOI: 10.3389/fgene.2025.1541541
The mechanism of RNA methylation writing protein-related prognostic genes in lung adenocarcinoma based on bioinformatics
Abstract
Objective: RNA methylation modifications play biological roles in tumorigenicity and immune response, mainly mediated by the "writer" enzyme. Lung adenocarcinoma (LUAD) development is closely related to RNA methylation. Here, the prognostic values of the "writer" enzymes and the tumor immunosurveillance in LUAD aim to provide new theoretical references for the research of LUAD.
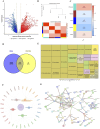
Methods: Genes associated with RNA methylation writer protein in LUAD were identified using The Cancer Genome Atlas Program (TCGA) data and weighted gene co-expression network analysis (WGCNA). Independent prognostic factors were screened by Cox regression and least absolute shrinkage and selection operator (LASSO) regression analyses. A prognostic risk model and a nomogram were established using these genes. Moreover, Gene Set Enrichment Analysis (GSEA) and CIBERSORTx were used to analyze the immune cell infiltration and enrichment pathways in the low- and high-risk groups, respectively. In addition, genes' potential functions and regulatory mechanisms were explored through gene-gene interaction (GGI) networks and competing endogenous RNA (ceRNA) networks.
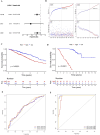
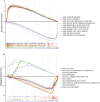
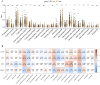
Results: We selected 202 genes associated with RNA methylation writer proteins, from which we identified the three genes (CLEC3B, GRIA1, and ANOS1). A prognostic risk model was constructed based on genes associated with RNA methylation writer proteins and stage, demonstrating reliable predictive performance. GGI analysis revealed GRIA1 as a crucial gene. Enrichment analysis revealed that the high-risk group had upregulated pathways connected to cell division. Additionally, immune infiltration analysis revealed that the significantly higher levels of NK cells, activated mast cells, activated CD4 memory cells, and M0 and M1 macrophages displayed in the high-risk group, while the significantly lower levels of monocytes, dendritic cells, M2 macrophages, and inactive CD4 memory cells were in the low-risk group. Moreover, Spearman correlation analysis demonstrated that the three prognostic genes and risk scores correlated highly with various immune cells.
Conclusion: This study identified three prognostic genes related to RNA methylation writer proteins in LUAD. A reliable prognostic model was constructed. The identified prognostic genes also play significant roles in immune cell infiltration in LUAD. This study provides new theoretical references for subsequent in-depth research on LUAD.
Keywords: RNA methylation writer proteins; biomarker; immune cell infiltration; lung adenocarcinoma; prognostic risk mode.
Copyright © 2025 Yin, Luo and Luo.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Similar articles
-
Molecular subtypes of lung adenocarcinoma patients for prognosis and therapeutic response prediction with machine learning on 13 programmed cell death patterns.J Cancer Res Clin Oncol. 2023 Oct;149(13):11351-11368. doi: 10.1007/s00432-023-05000-w. Epub 2023 Jun 28. J Cancer Res Clin Oncol. 2023. PMID: 37378675 Free PMC article.
-
Construction of a prognostic model for lung adenocarcinoma based on necroptosis genes and its exploration of the potential for tumor immunotherapy.Transl Cancer Res. 2025 May 30;14(5):2563-2579. doi: 10.21037/tcr-24-2165. Epub 2025 May 26. Transl Cancer Res. 2025. PMID: 40530113 Free PMC article.
-
Bioinformatics analysis of BTK expression in lung adenocarcinoma: implications for immune infiltration, prognostic biomarkers, and therapeutic targeting.3 Biotech. 2024 Sep;14(9):215. doi: 10.1007/s13205-024-04053-z. Epub 2024 Aug 28. 3 Biotech. 2024. PMID: 39220827
-
Taurine up-regulated 1: A dual regulator in immune cell-mediated pathogenesis of human diseases.Prog Biophys Mol Biol. 2025 Jul 15;197:84-96. doi: 10.1016/j.pbiomolbio.2025.07.002. Online ahead of print. Prog Biophys Mol Biol. 2025. PMID: 40675392 Review.
-
m5C RNA methylation in cancer: from biological mechanism to clinical perspectives.Eur J Med Res. 2025 Jun 21;30(1):503. doi: 10.1186/s40001-025-02812-z. Eur J Med Res. 2025. PMID: 40544318 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources
Research Materials

