Proteo-metabolomic analysis of fruits reveals molecular insights into variations among Italian Sweet Cherry (Prunus avium L.) accessions
- PMID: 40530268
- PMCID: PMC12170513
- DOI: 10.3389/fpls.2025.1591996
Proteo-metabolomic analysis of fruits reveals molecular insights into variations among Italian Sweet Cherry (Prunus avium L.) accessions
Abstract
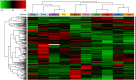
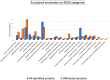
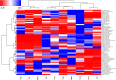
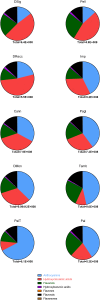
Mass spectrometry-based proteomics and metabolomics tackle the complex interactions between proteins and metabolites in fruits. Independently used to discern phenotypic disparities among plant accessions, these analytical approaches complement well-established DNA fingerprinting methods for assessing genetic variability and hereditary distance. To verify the applicability of integrated proteomic and metabolomic procedures in evaluating phenotypic differences between sweet cherry cultivars, and to potentially relate these findings to specific pomological traits, we conducted a comparative analysis of fruits from ten Italian accessions. We identified 3786 proteins, of which 288 exhibited differential representation between ecotypes, including key components influencing fruit quality and allergenic potential. Furthermore, 64 polyphenols were identified, encompassing anthocyanins, hydroxycinnamic acids, flavanols, hydroxybenzoic acids, flavonols, and flavanones subgroups. Multivariate analysis of total quantitative data outlined cultivar differences and phenotypic relationships. Coherent associations between proteomic and metabolomic data underscored their complementary role in characterizing genetic relationships elucidated through DNA fingerprinting techniques. Proteo-metabolomic results verified a certain correlation between the relative abundance of specific polyphenols, enzymes involved in their metabolism, and color characteristics of fruits. These findings highlight the significance of integrating results from diverse omics approaches to reveal molecular drivers of ecotype-specific traits and identify biomarkers for selecting and breeding cultivars in the next future.
Keywords: Prunus avium; biodiversity; fruit; metabolomics; proteomics; sweet cherry.
Copyright © 2025 De Pascale, Troise, Petriccione, Nunziata, Cice, Ferrara, Scaloni and Salzano.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Figures

Similar articles
-
Plastomic evolution and genetic diversity of cultivated sweet cheery (Prunus avium (L.) L.) in China.BMC Plant Biol. 2025 Aug 8;25(1):1043. doi: 10.1186/s12870-025-07125-1. BMC Plant Biol. 2025. PMID: 40781270 Free PMC article.
-
Investigating phenotypic relationships in persimmon accessions through integrated proteomic and metabolomic analysis of corresponding fruits.Front Plant Sci. 2023 Jan 30;14:1093074. doi: 10.3389/fpls.2023.1093074. eCollection 2023. Front Plant Sci. 2023. PMID: 36794209 Free PMC article.
-
Integrating Gut Microbiome and Metabolomics with Magnetic Resonance Enterography to Advance Bowel Damage Prediction in Crohn's Disease.J Inflamm Res. 2025 Jun 11;18:7631-7649. doi: 10.2147/JIR.S524671. eCollection 2025. J Inflamm Res. 2025. PMID: 40535353 Free PMC article.
-
A meta-analysis of genome-wide association studies to identify candidate genes associated with feed efficiency traits in pigs.J Anim Sci. 2025 Jan 4;103:skaf010. doi: 10.1093/jas/skaf010. J Anim Sci. 2025. PMID: 39847436 Free PMC article.
-
Defining disease severity in atopic dermatitis and psoriasis for the application to biomarker research: an interdisciplinary perspective.Br J Dermatol. 2024 Jun 20;191(1):14-23. doi: 10.1093/bjd/ljae080. Br J Dermatol. 2024. PMID: 38419411 Free PMC article. Review.
References
-
- Blando F., Oomah B. D. (2019). Sweet and sour cherries: Origin, distribution, nutritional composition and health benefits. Trends Food Sci. Technol. 86, 517. doi: 10.1016/j.tifs.2019.02.052 - DOI
LinkOut - more resources
Full Text Sources

