A long non-coding RNA lncRNA18313 regulates resistance against cadmium stress in wheat
- PMID: 40530269
- PMCID: PMC12171198
- DOI: 10.3389/fpls.2025.1583758
A long non-coding RNA lncRNA18313 regulates resistance against cadmium stress in wheat
Abstract
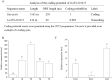
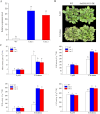
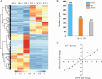
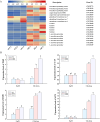
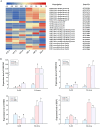
Long non-coding RNAs (lncRNAs) have been demonstrated to play key roles in plant response and adaptation to heavy metal stresses. However, the exact biological functions and potential regulatory mechanism, especially in wheat's response to cadmium (Cd) stress, are still poorly understood. We have previously discovered a Cd stress-related lncRNA in wheat, namely TalncRNA18313. In this study, qRT-PCR analysis revealed that TalncRNA18313 was expressed extensively in wheat leaves, and its accumulation was highly induced by Cd stress. To further fully explore the function of lncRNA18313 in response to Cd stress, lncRNA18313 was cloned from wheat (Triticum aestivum L.), and was transformed into Arabidopsis. When TalncRNA18313 was heterologous expressed in Arabidopsis, the transgenic plants exhibited enhanced Cd tolerance characterized by lower malondialdehyde (MDA) levels and higher activities of key antioxidant enzymes, such as catalase (CAT), superoxide dismutase (SOD) and peroxidase (POD). Subsequently, RNA-sequencing (RNA-seq) analysis demonstrated that 370 genes were differentially expressed in lncRNA18313 overexpressing transgenic lines under Cd stress comparing to wild type plants. Among the genes regulated by lncRNA18313, the most significantly enriched were those involved in transcriptional regulation and antioxidative defense responses. These results suggest that TalncRNA18313 plays a crucial role in improving Cd tolerance in wheat by modulating key stress-related pathways, particularly those critical for coping with oxidative damage and regulating gene expression under Cd stress. This discovery contributes to the expanding understanding of knowledge about the involvement of lncRNAs in plant stress responses and offers promising potential for improving crop resilience to environmental stresses.
Keywords: Cd stress; RNA-Seq; heavy metal; long non-coding RNA; wheat.
Copyright © 2025 Zhao, Bai, Fan, Zhu and Qiu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Overexpression of a Malus baccata (L.) Borkh WRKY Factor Gene MbWRKY33 Increased High Salinity Stress Tolerance in Arabidopsis thaliana.Int J Mol Sci. 2025 Jun 18;26(12):5833. doi: 10.3390/ijms26125833. Int J Mol Sci. 2025. PMID: 40565296 Free PMC article.
-
Salicylic acid confers cadmium tolerance in wheat by regulating photosynthesis, yield and ionic homeostasis.Sci Rep. 2025 Jan 29;15(1):3698. doi: 10.1038/s41598-025-87236-9. Sci Rep. 2025. PMID: 39880835 Free PMC article.
-
An in - silico perspective on the role of methylation-related genes in wheat - Fusarium graminearum interaction.3 Biotech. 2025 Jan;15(1):12. doi: 10.1007/s13205-024-04179-0. Epub 2024 Dec 16. 3 Biotech. 2025. PMID: 39698303
-
LncRNAOmics: A Comprehensive Review of Long Non-Coding RNAs in Plants.Genes (Basel). 2025 Jun 29;16(7):765. doi: 10.3390/genes16070765. Genes (Basel). 2025. PMID: 40725421 Free PMC article. Review.
-
Assessing the comparative effects of interventions in COPD: a tutorial on network meta-analysis for clinicians.Respir Res. 2024 Dec 21;25(1):438. doi: 10.1186/s12931-024-03056-x. Respir Res. 2024. PMID: 39709425 Free PMC article. Review.
References
-
- Bai Y., He J., Yao Y., An L., Cui Y., Li X., et al. (2024). Identification and functional analysis of long non−coding RNA (lncRNA) and metabolites response to mowing in hulless barley (Hordeum vulgare L. var. nudum hook. f.). BMC Plant Biol. 24, 666. doi: 10.1186/s12870-024-05334-8 - DOI - PMC - PubMed
-
- Cao Y., Du P., Zhang J., Ji J., Xu J., Liang B. (2023). Dopamine alleviates cadmium stress in apple trees by recruiting beneficial microorganisms to enhance the physiological resilience revealed by high-throughput sequencing and soil metabolomics. Hortic. Res. 10, 112. doi: 10.1093/hr/uhad112 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

