The sucrose synthase gene family in blueberry (Vaccinium darrowii): functional insights into the role of VdSUS4 in salt stress tolerance
- PMID: 40530277
- PMCID: PMC12171176
- DOI: 10.3389/fpls.2025.1581182
The sucrose synthase gene family in blueberry (Vaccinium darrowii): functional insights into the role of VdSUS4 in salt stress tolerance
Abstract
Introduction: The sucrose synthase (SUS), a crucial enzyme in the sucrose metabolism, is encoded by a multigene family in plant kingdom.
Methods: In our study, we utilized bioinformatics tools to identify and characterize the members of the SUS gene family within the blueberry genome. Our analysis encompassed the physicochemical properties, gene structures, conserved motifs, promoter cis-acting elements, chromosomal locations, evolutionary relationships and expression profiles of these family members, allowing us to predict their potential functions.
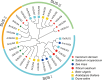
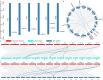
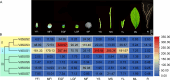
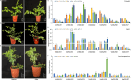
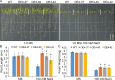
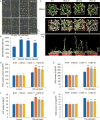
Results: We identified seven distinct SUS genes, mapped across six chromosomes, showcasing the complexity of this gene family in blueberries. Phylogenetic analysis, constructed through a multi-species phylogenetic tree, revealed that the SUS gene family can be categorized into three subfamilies: SUS I, SUS II and SUS III. Notable variations were observed among the VdSUS gene family members, particularly in the number of amino acids, molecular weight, isoelectric point, and hydrophobicity of the encoded proteins. Intriguingly, our predictive analysis of the promoter regions of VdSUS genes uncovered a wealth of cis-acting elements linked to light response, hormonal regulation, and stress responses, suggesting a role in adaptive mechanisms. Expression studies indicated that VdSUS genes were highly expressed in fruit tissues, with the application of exogenous sucrose leading to significant downregulation of VdSUS2, VdSUS3 and VdSUS6. Furthermore, the expression of VdSUS genes was found to be responsive to abiotic stresses, such as salt, drought, and low temperatures, with varying degrees of upregulation or downregulation observed. Most notably, the overexpression of VdSUS4 in Arabidopsis thaliana resulted in enhanced tolerance to salt stress.
Discussion: These findings have shed new light on the multifaceted roles of VdSUS gene family members in the complex physiological processes of blueberries, highlighting their potential in the context of stress adaptation and fruit development.
Keywords: bioinformatics; blueberry; salt stress; sucrose synthase; tissue-specific expression.
Copyright © 2025 Wang, Yang, Geng, Zhang and Zhou.
Conflict of interest statement
Authors LY, HXZ, and HJZ were employed by Bestplant Shandong Stem Cell Engineering Co., Ltd. Authors LY, HXZ, and HJZ were employed by Zhaoyuan Shenghui Agricultural Technology Development Co., Ltd. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Baroja-Fernández E., Muñoz F. J., Montero M., Etxeberria E., Sesma M. T., Ovecka M., et al. (2009). Enhancing sucrose synthase activity in transgenic potato (Solanum tuberosum L.). tubers results in increased levels of starch., ADPglucose and UDPglucose and total yield. Plant Cell Physiol. 50, 1651–1662. doi: 10.1093/pcp/pcp108 - DOI - PubMed
LinkOut - more resources
Full Text Sources

