Integrated transcriptomic and metabolomic analyses provide new insights into alkaline stress tolerance in Gossypium hirsutum
- PMID: 40530291
- PMCID: PMC12170613
- DOI: 10.3389/fpls.2025.1604606
Integrated transcriptomic and metabolomic analyses provide new insights into alkaline stress tolerance in Gossypium hirsutum
Abstract
Introduction: Cotton, one of the most important economic crops worldwide, has long been bred mainly for improvements in yield and quality, with relatively little focus on salt-alkali resistance.
Methods: In this study, transcriptomic and metabolomic sequencing were performed on Gossypium hirsutum exposed to alkaline stress for different durations.
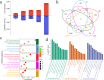
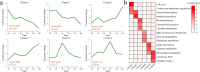
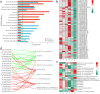
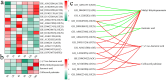
Results: The results of sample clustering, principal component analysis (PCA), and the number of differentially expressed genes (DEGs) revealed that 12 hours and 24 hours were the periods during which upland cotton presented the strongest response to salt stress, with flavonoid biosynthesis and alpha-linolenic acid metabolism playing significant roles during this time. A total of 6,610 DEGs were identified via comparison to the 0 h time point, including 579 transcription factors (TFs) that were significantly enriched in pathways such as flavonoid biosynthesis, the cell cycle, the cytochrome P450 pathway, phenylalanine metabolism, phototransduction, and alpha-linolenic acid metabolism. Through ultrahigh-performance liquid chromatography-MS (UPLC-MS), 4,225 metabolites were identified, and 1,684 differentially accumulated metabolites (DAMs) were identified by comparison to the levels at 0 h. A joint analysis of RNA-seq and metabolomic data revealed that the flavonoid biosynthesis and alpha-linolenic acid metabolism pathways play key roles in the response of G. hirsutum to alkaline stress, and the key genes in these pathways were identified. The weighted gene correlation network analysis (WGCNA) revealed 15 candidate genes associated with alkali tolerance in cotton, including 4 TFs and 4 genes related to flavonoid and anthocyanin biosynthesis.
Conclusion: In conclusion, our study provides a theoretical foundation for understanding the molecular mechanisms underlying alkali tolerance in cotton and offers new gene resources for future research.
Keywords: Gossypium hirsutum; RNA-seq; alkaline stress; candidate genes; metabolome.
Copyright © 2025 Geng, Gao, Sun, Yang, Ma, Wang, Wang, Wang, Qian, Li and Zhao.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




Similar articles
-
Metabolomic and transcriptomic analyses of drought resistance mechanisms in sorghum varieties.PeerJ. 2025 Jul 4;13:e19596. doi: 10.7717/peerj.19596. eCollection 2025. PeerJ. 2025. PMID: 40625922 Free PMC article.
-
Physiological and broadly targeted metabolomic analyses of barley (Hordeum vulgare L.) in response to low-temperature stress.BMC Genomics. 2025 Jul 1;26(1):618. doi: 10.1186/s12864-025-11516-x. BMC Genomics. 2025. PMID: 40597568 Free PMC article.
-
Gh_FBL43 regulates the resistance of Gossypium hirsutum to Verticillium wilt through jasmonic acid and flavonoid-related pathways.Front Plant Sci. 2025 Jul 17;16:1620947. doi: 10.3389/fpls.2025.1620947. eCollection 2025. Front Plant Sci. 2025. PMID: 40747528 Free PMC article.
-
A meta-analysis of genome-wide association studies to identify candidate genes associated with feed efficiency traits in pigs.J Anim Sci. 2025 Jan 4;103:skaf010. doi: 10.1093/jas/skaf010. J Anim Sci. 2025. PMID: 39847436 Free PMC article.
-
Defining disease severity in atopic dermatitis and psoriasis for the application to biomarker research: an interdisciplinary perspective.Br J Dermatol. 2024 Jun 20;191(1):14-23. doi: 10.1093/bjd/ljae080. Br J Dermatol. 2024. PMID: 38419411 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources

