Unraveling the mitochondrial genome of the medicinal Chinese motherwort (Leonurus japonicus, Lamiaceae): structural dynamics, organelle-to-nuclear gene transfer, and evolutionary implications
- PMID: 40530293
- PMCID: PMC12170669
- DOI: 10.3389/fpls.2025.1546449
Unraveling the mitochondrial genome of the medicinal Chinese motherwort (Leonurus japonicus, Lamiaceae): structural dynamics, organelle-to-nuclear gene transfer, and evolutionary implications
Abstract
Introduction: Leonurus japonicus (Chinese motherwort) is a medicinal Lamiaceae species renowned for its pharmacological compounds, yet its mitochondrial genome remains unexplored. Elucidating mitogenomic structure and evolution can inform plant genetics, phylogenetics, and molecular breeding.
Methods: We assembled the complete mitochondrial genome of L. japonicus using a combination of Oxford Nanopore long reads and Illumina short reads. Three assembly strategies-de novo assembly with PMAT and Flye, and hybrid assembly with Unicycler-were integrated and validated via read mapping and comparison to reference mitogenomes (Salvia miltiorrhiza, Arabidopsis thaliana, Liriodendron tulipifera). Annotation employed GeSeq, tRNAscan-SE, and manual curation. Repeat elements (SSR, tandem, dispersed) were identified with MISA, TRF, and REPuter; plastid-to-mitochondrion transfers (MTPTs) were detected by BLASTN against the assembled plastome; and RNA editing sites were predicted using Deepred-mt. Phylogenetic and synteny analyses were conducted with IQ-TREE, MAFFT alignments of 24 conserved PCGs, and NGenomeSyn visualization.
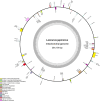
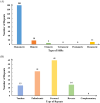
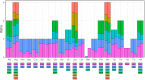
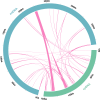
Results: The circular mitogenome spanned 384,199 bp (45.1% GC) and encoded 35 protein-coding genes, 11 tRNAs, and 3 rRNAs. We detected 241 SSRs, 13 tandem repeats, and 90 dispersed repeats, indicating extensive recombination potential. Thirty-one MTPTs totaling 24,818 bp (6.46% of the mitogenome) were identified. Comparative analyses revealed strong purifying selection (Ka/Ks < 1) across most PCGs, with selective signatures in atp4 and ccmB. Phylogenetic inference placed L. japonicus among Lamiales, closely allied to Scutellaria tsinyunensis and Rotheca serrata. Synteny maps demonstrated frequent genome rearrangements. Deepred-mt predicted 408 C-to-U RNA editing sites, notably in nad4 and ccmB, including novel start and stop codons.
Discussion: The L. japonicus mitogenome exhibits marked structural plasticity, reflecting dynamic repeats and organelle-to-organelle DNA transfers. Extensive RNA editing underscores post-transcriptional regulation in mitochondrial function. These findings enrich genomic resources for Leonurus, support phylogenetic and evolutionary studies in Lamiaceae, and lay groundwork for molecular breeding and conservation strategies targeting mitochondrial traits.
Keywords: Leonurus japonicus; RNA editing; long-read sequencing; mitochondrial genome; phylogenetic relationships; plastid-derived DNA; repeat sequence; synteny analysis.
Copyright © 2025 Bai, Zhu, Chen, Wang, Liu, Feng, Huang, Lee, Kokubugata, Qi and Yan.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
LinkOut - more resources
Full Text Sources
Miscellaneous

