Gonadotropins differentially regulate testicular cell adhesion and junctional complexes during flatfish spermiogenesis through the oxytocin and relaxin signaling pathways
- PMID: 40530338
- PMCID: PMC12171224
- DOI: 10.3389/fcell.2025.1574690
Gonadotropins differentially regulate testicular cell adhesion and junctional complexes during flatfish spermiogenesis through the oxytocin and relaxin signaling pathways
Abstract
Introduction: The molecular mechanisms regulating teleost semicystic spermatogenesis remain largely unknown. In the flatfish Senegalese sole (Solea senegalensis), haploid round spermatids released into the lumen of the seminiferous tubules resume spermiogenesis (the differentiation of germ cells into spermatozoa) in response to the luteinizing hormone (Lh). However, how the spermatids detach from Sertoli cells and how Lh crosses the blood-testis barrier (BTB) are yet to be determined.
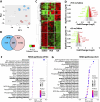
Methods: Here, we used an RNA-seq transcriptomic analysis of the testis from sole males treated with recombinant follicle stimulating hormone and Lh (rFsh and rLh, respectively).
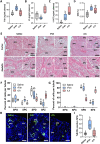
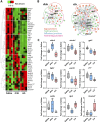
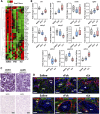
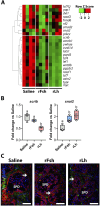
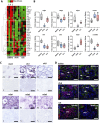
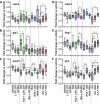
Results: This analysis reveals that both gonadotropins differentially downregulate a number of transcripts potentially encoding cell-cell junction and adhesion proteins, as well as components of the Oxytocin (Oxt) and Relaxin (Rln) signaling pathways. In situ hybrizidation and immunolocalization experiments confirmed the formation of adherens, gap, and tight junctions between Sertoli cells, and between Sertoli cells and spermatids. Using these methods, we also verified the expression of Oxt and Rln peptides and their cognate receptors in these cells. Further in vitro assays using testicular explants incubated with Oxt, Rln and inhibitors of their receptors, combined with rFsh or rLh, showed that the gonadotropic-induced transcriptional repression of cell junction and adhesion genes in the seminiferous epithelium, particularly by Lh, was largely mediated by the downregulation of Oxt and Rln signaling.
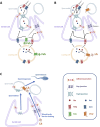
Discussion: These data suggest that the Oxt- and Rln-mediated gonadotropic disruption of the BTB and Sertoli cells-spermatid junctions in the sole testis facilitates spermatid release and Lh paracellular transport into the seminiferous lumen during spermiogenesis.
Keywords: adherens junctions; blood-testis barrier; cell junctions; endocrine control; spermatogenesis; teleost.
Copyright © 2025 López-Fortún, Roig-Genovés, Giménez, Cerdà and Chauvigné.
Conflict of interest statement
Authors JR-G and IG were employed by Rara Avis Biotec, S.L. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
LinkOut - more resources
Full Text Sources
Miscellaneous

