Cytotoxic NK Cells Impede Response to Checkpoint Immunotherapy in Melanoma with an Immune-Excluded Phenotype
- PMID: 40530504
- PMCID: PMC12409281
- DOI: 10.1158/2159-8290.CD-24-1208
Cytotoxic NK Cells Impede Response to Checkpoint Immunotherapy in Melanoma with an Immune-Excluded Phenotype
Abstract
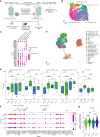
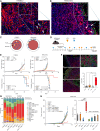
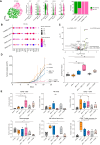
Immune checkpoint blockade (ICB) has revolutionized cancer treatment. Unfortunately, the inability of lymphocytes to infiltrate the tumor nest, a phenomenon known as immune exclusion, drastically limits ICB responsiveness. Analyzing the immune landscape of matched pre- and early on-treatment biopsies of patients with melanoma undergoing ICB therapy, we observed a significant increase in cytotoxic NK cells in early on-treatment biopsies from nonresponders. Spatial multiomic analyses revealed that, although NK cells colocalized with CD8+ T cells within the tumor bed in responding lesions, they were excluded from the tumor parenchyma in nonresponding lesions. Strikingly, pharmacologic depletion of NK cells in a unique melanoma mouse model exhibiting an immune-excluded phenotype unleashed immune infiltration of the tumor core and tumor clearance upon ICB exposure. Mechanistically, we show that NK cells are actively recruited to immune-excluded areas upon ICB exposure via the chemokine receptor CX3CR1 to suppress tumor infiltration and antitumor function of CD8+ T cells.
Significance: Immune exclusion is responsible for intrinsic resistance to ICB in about half of nonresponder patients. Our unexpected observation that targeting NK cell biology unleashes the recruitment and antitumor activity of CD8+ T cells in tumors with an immune-excluded phenotype offers a potential therapeutic avenue for this large patient population. See related commentary by Galvez-Cancino et al., p. 1777 See related article by Song et al., p. 1835.
©2025 The Authors; Published by the American Association for Cancer Research.
Conflict of interest statement
J. Pozniak reports personal fees from Stichting Tegen Kanker and H2020-MSCA-IF-2019 during the conduct of the study. N. Roda reports personal fees from Fonds Wetenschappelijk Onderzoek during the conduct of the study. A. De Visscher reports grants from Fonds Wetenschappelijk Onderzoek Vlaanderen during the conduct of the study. P.G. Demaerel reports personal fees from Fonds Wetenschappelijk Onderzoek during the conduct of the study. J. Declercq reports grants from Fonds Wetenschappelijk Onderzoek, VIB vzw, Stichting Tegen Kanker, and iBOF during the conduct of the study. C. Gkemisis reports personal fees from Fonds Wetenschappelijk Onderzoek during the conduct of the study. L. Pollaris reports grants from Fonds Wetenschappelijk Onderzoek Vlaanderen during the conduct of the study. F.M. Bosisio reports grants from Fonds Wetenschappelijk Onderzoek Fundamenteel Klinisch Mandaat EMH-D8972-FKM/20 during the conduct of the study. Z. Li reports personal fees from HanchorBio and Henlius and other support from Alphamab Oncology outside the submitted work. No disclosures were reported by the other authors.
Figures

References
-
- Cursons J, Souza-Fonseca-Guimaraes F, Foroutan M, Anderson A, Hollande F, Hediyeh-Zadeh S, et al. A gene signature predicting natural killer cell infiltration and improved survival in melanoma patients. Cancer Immunol Res 2019;7:1162–74. - PubMed
MeSH terms
Substances
Grants and funding
- #896897/HORIZON EUROPE Marie Sklodowska-Curie Actions (MSCA)
- 2023/2310/Stichting Tegen Kanker (Fondation Contre le Cancer)
- 2022-178/Stichting Tegen Kanker (Fondation Contre le Cancer)
- 2022-037/Stichting Tegen Kanker (Fondation Contre le Cancer)
- 2024-198/Stichting Tegen Kanker (Fondation Contre le Cancer)
- 1253225N-7028/Fonds Wetenschappelijk Onderzoek (FWO)
- 12ATN24N/Fonds Wetenschappelijk Onderzoek (FWO)
- EMH-D8191-AKUL/19/30 I005920N/KU Leuven (Katholieke Universiteit Leuven)
- EMH-D8972-FKM/20/Fonds Wetenschappelijk Onderzoek (FWO)
- 11J7324N/The Research Foundation Flanders
- 11K0722N/Fonds Wetenschappelijk Onderzoek (FWO)
- G0B1622N/FWO/KOTK
- G070622N/FWO/KOTK
- G045824N/FWO/KOTK
- S006825N/FWO-SBO
- #23/005/iBOF
- EOS/FWO/FRS-FNRS
- # 40007513/FWO/FRS-FNRS
LinkOut - more resources
Full Text Sources
Medical
Research Materials

