Next generation genetic screens in kinetoplastids
- PMID: 40530689
- PMCID: PMC12203798
- DOI: 10.1093/nar/gkaf515
Next generation genetic screens in kinetoplastids
Abstract
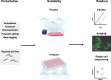
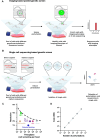
The genomes of all organisms encode diverse functional elements, including thousands of genes and essential noncoding regions for gene regulation and genome organization. Systematic perturbation of these elements is crucial to understanding their roles and how their disruption impacts cellular function. Genetic perturbation approaches, which disrupt gene expression or function, provide valuable insights by linking genetic changes to observable phenotypes. However, perturbing individual genomic elements one at a time is impractical. Genetic screens overcome this limitation by enabling the simultaneous perturbation of numerous genomic elements within a single experiment. Traditionally, these screens relied on simple, high-throughput readouts such as cell fitness, differentiation, or one-dimensional fluorescence. However, recent advancements have introduced powerful technologies that combine genetic screens with image-based and single-cell sequencing readouts, allowing researchers to study how perturbations affect complex cellular phenotypes on a genome-wide scale. These innovations, alongside the development of CRISPR-Cas technologies, have significantly enhanced the precision, efficiency, and scalability of genetic screening approaches. In this review, we discuss the genetic screens performed in kinetoplastid parasites to date, emphasizing their application to both coding and noncoding regions of the genome. Furthermore, we explore how integrating image-based and single-cell sequencing technologies with genetic screens holds the potential to deliver unprecedented insights into cellular function and regulatory mechanisms.
© The Author(s) 2025. Published by Oxford University Press on behalf of Nucleic Acids Research.
Conflict of interest statement
No conflicts of interest declared.
Figures



References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

