Non-coding Y RNA fragments in a complex with YBX1 modulate PARP1 residency at DNA double strand breaks
- PMID: 40530699
- PMCID: PMC12203914
- DOI: 10.1093/nar/gkaf517
Non-coding Y RNA fragments in a complex with YBX1 modulate PARP1 residency at DNA double strand breaks
Abstract
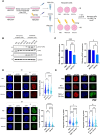
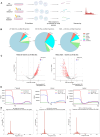
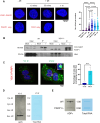
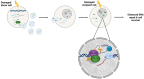
To protect genome integrity from pervasive threats of damage and prevent diseases like cancer, cells employ an integrated network of signalling pathways called the DNA damage response. These pathways involve both protein and RNA components, which can act within the damaged cell or be transferred intercellularly to influence population-wide responses to damage. Here, we show that radioprotection can be conferred by damage-derived exosomes and is dependent on YBX1-packaged Y3-derived ysRNA. In recipient cells, ysRNAs are methylated on cytosine and bound by m5C reader, YBX1. YBX1/ysRNA localise at double strand break (DSB) sites to promote efficient DNA repair and cell survival through complex formation with PARP1. YBX1 modulates PARP1 auto-modification by facilitating ysRNA ADP-ribosylation, promoting increased PARP1 residency at DSBs. Our data highlight an unprecedented role for these under-studied species of small non-coding RNAs, identifying them as a novel substrate for PARP1 mediated ADP-ribosylation with a function in DNA repair.
© The Author(s) 2025. Published by Oxford University Press on behalf of Nucleic Acids Research.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

