Comparative and functional genomic analysis of foreign DNA defense mechanisms in Enterococcus faecium
- PMID: 40530811
- PMCID: PMC12323599
- DOI: 10.1128/spectrum.00289-25
Comparative and functional genomic analysis of foreign DNA defense mechanisms in Enterococcus faecium
Abstract
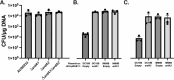
Enterococcus faecium is notoriously difficult to study genetically due to the poor understanding of barriers preventing foreign DNA uptake, such as the proteins that modify type I restriction modification (RM) system activity. Here, we compared E. faecium repertoires of the HsdS specificity subunit (dictating the DNA motif that is adenine methylated) from type I RM systems among 805 globally reported E. faecium isolates. We showed there were eight distinct HsdS types, with four dominant variants that were also significantly enriched in the hospital-associated clade A1 E. faecium lineage. Adenine methylome analysis of a subset of eight representative E. faecium strains revealed that only two exhibited functional type I RM systems, with the activity corroborated by the construction of type I RM deletion mutants. To investigate this surprising finding, we assessed the contribution of the anti-restriction protein ArdA that specifically inhibits type I RM function. The E. faecium ST796 clinical isolate AUS0233 has one intact type I RM system, no adenine methylation, and two distinct ardA paralogs. When heterologously expressed in Staphylococcus aureus JE2, both E. faecium ardA variants were functional, each inhibiting the function of the two type I RM systems in S. aureus. However, the deletion of one or both versions of ardA in E. faecium AUS0233 did not change the transformation efficiency with exogenous DNA, suggesting ArdA in E. faecium AUS0233 is not controlling type I RM. This study highlights the complexity of DNA defense mechanisms in E. faecium and suggests that unidentified factors control the acquisition of foreign DNA.IMPORTANCEEnterococcus faecium has mechanisms of DNA methylation and targeted DNA degradation (called restriction modification [RM]) that hinder foreign DNA uptake, thus influencing the acquisition of important phenotypes such as antibiotic resistance. Restriction barriers also frustrate efforts for laboratory genetic manipulation used to study this pathogen. From PacBio analysis of E. faecium strains, it was observed that the majority of E. faecium do not adenine methylate DNA despite genome analysis indicating they have intact type I RM methylation systems. One explanation for this observation is that E. faecium produces anti-restriction factors such as ArdA, which can inhibit type I RM systems. However, the deletion of both ardA alleles did not improve the efficiency of DNA uptake. These findings build our foundational knowledge of how E. faecium controls foreign DNA and show there is additional complexity surrounding these systems to be discovered.
Keywords: DNA defense mechanisms; Enterococcus faecium; genomics; hsdS; restriction modification system.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
The Black Book of Psychotropic Dosing and Monitoring.Psychopharmacol Bull. 2024 Jul 8;54(3):8-59. Psychopharmacol Bull. 2024. PMID: 38993656 Free PMC article. Review.
-
Measures implemented in the school setting to contain the COVID-19 pandemic.Cochrane Database Syst Rev. 2022 Jan 17;1(1):CD015029. doi: 10.1002/14651858.CD015029. Cochrane Database Syst Rev. 2022. Update in: Cochrane Database Syst Rev. 2024 May 2;5:CD015029. doi: 10.1002/14651858.CD015029.pub2. PMID: 35037252 Free PMC article. Updated.
-
Systemic treatments for metastatic cutaneous melanoma.Cochrane Database Syst Rev. 2018 Feb 6;2(2):CD011123. doi: 10.1002/14651858.CD011123.pub2. Cochrane Database Syst Rev. 2018. PMID: 29405038 Free PMC article.
-
Short-Term Memory Impairment.2024 Jun 8. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2024 Jun 8. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 31424720 Free Books & Documents.
-
Signs and symptoms to determine if a patient presenting in primary care or hospital outpatient settings has COVID-19.Cochrane Database Syst Rev. 2022 May 20;5(5):CD013665. doi: 10.1002/14651858.CD013665.pub3. Cochrane Database Syst Rev. 2022. PMID: 35593186 Free PMC article.
References
-
- Fiore E, Tyne D, Gilmore MS. 2019. Pathogenicity of enterococci. In Gram-Positive Pathogens Third Edition, 3rd ed
-
- Kristich CJ, Rice LB, Arias CA. 2014. Enterococcal infection-treatment and antibiotic resistance. In Gilmore MS, Clewell DB, Ike Y, Shankar N (ed), Enterococci: from commensals to leading causes of drug resistant infection. Boston. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

