Chlorobaculum tepidum outer membrane vesicles may transport biogenic elemental sulfur
- PMID: 40530856
- PMCID: PMC12285230
- DOI: 10.1128/aem.01019-25
Chlorobaculum tepidum outer membrane vesicles may transport biogenic elemental sulfur
Abstract
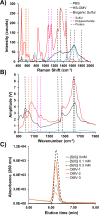
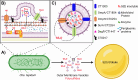
Outer membrane-derived vesicles (OMVs) have been studied in different phyla of gram-negative bacteria, most extensively in the Pseudomonadota, where they have been shown to participate in diverse biological and environmental processes. To date, the production of OMVs has not been reported in the Chlorobiaceae within the phylum Chlorobiota. Chlorobaculum tepidum is the model organism for the Chlorobiaceae that synthesizes and consumes insoluble extracellular sulfur (S(0)) globules by an unknown mechanism. Here, we report evidence implicating outer membrane vesicles in biogenic S(0) globule synthesis. We demonstrate that Cba. tepidum secretes OMVs in the extracellular milieu and that OMV concentration and size vary with growth conditions, particularly sulfide concentration. A core of 31 proteins involved in diverse biological processes such as cell wall biogenesis, inorganic ion transport, and metabolism was found to be shared between OMVs, extracellular S(0) globules, and Cba. tepidum-intact cells. Multiple analytical methods indicated that OMVs contain S(0) and that OMVs and biogenic S(0) globules share protein and polysaccharide signatures, including lipooligosaccharides. Together, these lines of evidence indicate that Cba. tepidum's OMVs are one component of sulfur transport between cells and extracellular sulfur globules.IMPORTANCEAll living cells must exchange material with their environment while maintaining cellular integrity. This is a particular challenge for materials that are not water-soluble; however, many bacteria utilize insoluble materials for energy conservation and as nutrients for growth. Here, we show that Cba. tepidum makes outer membrane vesicles, and these vesicles are likely involved in the exchange of material with extracellular elemental sulfur globules formed and consumed by Cba. tepidum as part of its energy metabolism based on oxidizing reduced sulfur compounds like hydrogen sulfide. These data expand our basic understanding of Cba. tepidum's metabolism. As elemental sulfur is an industrial by-product with a limited number of uses, the information here may help enable the use of additional sulfur compounds by Cba. tepidum to drive the synthesis of biomass and/or specialty biochemicals from waste elemental sulfur by this autotrophic bacterium.
Keywords: Chlorobaculum tepidum; Chlorobiaceae; NanoIR spectroscopy; Raman spectroscopy; outer membrane vesicles; proteomics; sulfur globules.
Conflict of interest statement
Y.Y. is a named inventor on a patent application (PCT/US2023/020215) for the E3 technology, which has been licensed exclusively to CDS Analytical LLC (Oxford, PA).
Figures





Update of
-
Chlorobaculum tepidum Outer Membrane Vesicles May Transport Biogenic Elemental Sulfur.bioRxiv [Preprint]. 2025 May 13:2024.09.30.615447. doi: 10.1101/2024.09.30.615447. bioRxiv. 2025. Update in: Appl Environ Microbiol. 2025 Jul 23;91(7):e0101925. doi: 10.1128/aem.01019-25. PMID: 40463023 Free PMC article. Updated. Preprint.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

