ARTP mutagenesis for genome-wide identification of genes important for biofilm regulation in spoilage bacterium Pseudomonas fluorescens PF08
- PMID: 40530873
- PMCID: PMC12175535
- DOI: 10.1128/aem.00218-25
ARTP mutagenesis for genome-wide identification of genes important for biofilm regulation in spoilage bacterium Pseudomonas fluorescens PF08
Abstract
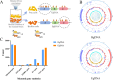
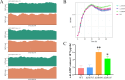
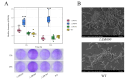
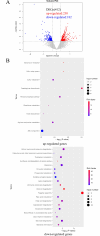
Pseudomonas fluorescens is a vital food spoilage bacterium and commonly spoils foods in the form of biofilms. Yet its biofilm regulation strategies have not been fully revealed. Here, we conducted a genome-wide screen of genes important for biofilm regulation using atmospheric and room temperature plasma mutagenesis together with the whole-genome resequencing technology. Three genes (D7M10_RS02105, D7M10_RS27690, and D7M10_RS25705) encoding GGDEF-EAL domain-containing proteins were found to have different mutation manifestations between biofilm cells and free cells. On direct testing, null mutants of D7M10_RS02105 and especially D7M10_RS27690 exhibited significantly elevated cyclic di-GMP (c-di-GMP) levels. Further studies indicated that a higher level of c-di-GMP caused by the null mutant of D7M10_RS27690 triggered cell growth, the production of siderophore and exopolysaccharide as well as autoaggregation, and hindered cell motility, all of which together promote biofilm formation. RNA-sequencing analysis revealed the transcription profile regulated by D7M10_RS27690, mostly including flagellar assembly and peptidoglycan biosynthesis pathways. Therein, the downregulated genes enriched in flagellar assembly were verified by qRT-PCR; the result of which was in agreement with the decreased cell motility.IMPORTANCEBiofilms formed by spoilage bacterium Pseudomonas fluorescens will bring about food quality and safety issues. In this study, we present the establishment of a genetic method and verified its reliability and efficiency for identifying genes associated with biofilm regulation. The genes we discovered offer new perspectives on the mechanisms of biofilm regulation in spoilage bacterium P. fluorescens. Moreover, the gene screen method based on atmospheric and room temperature plasma mutagenesis and whole-genome resequencing-coupled technology overcomes the labor-intensive issues caused by traditional methods and should generally be suitable for identifying genes associated with biofilm formation or dispersion in other bacteria.
Keywords: ARTP mutagenesis; Pseudomonas fluorescens; biofilm; c-di-GMP; siderophore.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Aswathanarayan JB, Vittal RR. 2014. Attachment and biofilm formation of Pseudomonas fluorescens PSD4 isolated from a dairy processing line. Food Sci Biotechnol 23:1903–1910. doi: 10.1007/s10068-014-0260-8 - DOI
-
- Mlipano CL, Alistair G, Michael L. 2018. Detection of proteolysis in milk by Pseudomonas fluorescens using urea PAGE method. J Food Stud 7:14. doi: 10.5296/jfs.v7i1.12019 - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

