Evolutionary trends in Bombella apis CRISPR-Cas systems
- PMID: 40530883
- PMCID: PMC12282179
- DOI: 10.1128/msystems.00166-25
Evolutionary trends in Bombella apis CRISPR-Cas systems
Abstract
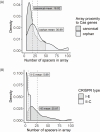
Bacteria and archaea employ a rudimentary immune system, CRISPR-Cas, to protect against foreign genetic elements such as bacteriophage. CRISPR-Cas systems are found in Bombella apis. B. apis is an important honey bee symbiont, found primarily in larvae, queens, and hive compartments. B. apis is found in the worker bee gut but is not considered a core member of the bee microbiome and has therefore been understudied with regard to its importance in the honey bee colony. However, B. apis appears to play beneficial roles in the colony, by protecting developing brood from fungal pathogens and by bolstering their development under nutritional stress. Previously, we identified CRISPR-Cas systems as being acquired by B. apis in its transition to bee association, as they are absent in a sister clade. Here, we assess the variation and distribution of CRISPR-Cas types across B. apis strains. We found multiple CRISPR-Cas types, some of which have multiple arrays, within the same B. apis genomes and also in the honey bee queen gut metagenomes. We analyzed the spacers between strains to identify the history of mobile element interaction for each B. apis strain. Finally, we predict interactions between viral sequences and CRISPR systems from different honey bee microbiome members. Our analyses show that the B. apis CRISPR-Cas systems are dynamic; that microbes in the same niche have unique spacers, which supports the functionality of these CRISPR-Cas systems; and that acquisition of new spacers may be occurring in multiple locations in the genome, allowing for a flexible antiviral arsenal for the microbe.
Importance: Honey bee worker gut microbes have been implicated in everything from protection from pathogens to breakdown of complex polysaccharides in the diet. However, there are multiple niches within a honey bee colony that host different groups of microbes, including the acetic acid bacterium Bombella apis. B. apis is found in the colony food stores, in association with brood, in worker hypopharyngeal glands, and in the queen's digestive tract. The roles that B. apis may serve in these environments are just beginning to be discovered and include the production of a potent antifungal that protects developing bees and supplementation of dietary lysine to young larvae, bolstering their nutrition. Niche specificity in B. apis may be affected by the pressures of bacteriophage and other mobile elements, which may target different strains in each specific bee environment. Studying the interplay between B. apis and its mobile genetic elements (MGEs) may help us better understand microbial community dynamics within the colony and the potential ramifications for the honey bee host.
Keywords: Bombella; CRISPR; Cas; defense systems; honey bee; microbiome; phage; royal jelly.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
- Anderson KE, Copeland DC. 2024. The honey bee “hive” microbiota: meta-analysis reveals a native and aerobic microbiota prevalent throughout the social resource niche. Front Bee Sci 2. doi: 10.3389/frbee.2024.1410331 - DOI
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

