High-throughput transcriptomics analysis of equipotent and human relevant mixtures of BPA alternatives reveal additive effects in vitro
- PMID: 40531193
- PMCID: PMC12408750
- DOI: 10.1007/s00204-025-04110-3
High-throughput transcriptomics analysis of equipotent and human relevant mixtures of BPA alternatives reveal additive effects in vitro
Abstract
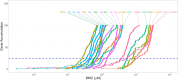
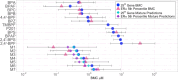
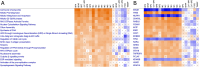
While many jurisdictions have phased out use of bisphenol A (BPA), there is increasing exposure to mixtures of BPA alternatives. Like BPA, some alternatives perturb nuclear hormone receptors and are endocrine disruptors. We used high-throughput transcriptomics (HTTr) to evaluate the potency and modes of action of seven mixtures of BPA alternatives and their 12 individual components in breast cancer cells. Our aim was to explore whether alternatives present in mixtures act additively. MCF-7 cells were exposed to chemicals (0.001-50 µM) for 48 h and gene expression analysis was used to measure global and estrogen receptor alpha (ERα)-specific transcriptomic changes. Transcriptomic points of departure (tPODs) were derived using benchmark concentration (BMC) modelling. We first identified concentrations at which global transcriptional activity was robustly altered. Then, we applied a ERα transcriptomic biomarker to identify ERα agonists and predict ERα activation tPODs. We employed mixtures modeling to predict potency of BPA alternatives and test for additive effects in vitro. Ingenuity pathway analysis (IPA; Qiagen) was used to identify upstream regulators and canonical pathways from genes fitting BMCs. BPAF was the most potent individual chemical tested overall, followed by BPA and BPC. All seven mixtures had additive effects across all tPODs modeled. The ERα transcriptomic biomarker classified all mixtures as ERα activators along with several BPA alternatives. All mixtures and most individual components perturbed similar upstream regulators and pathways, suggesting common modes of action. These data support the value of HTTr in identifying additive effects and toxicological potency of mixtures in vitro.
Keywords: Endocrine disruptor; Mixtures; Potency; Regulatory toxicology; Transcriptomics.
© 2025. Crown.
Conflict of interest statement
Conflict of Interest: The information in this document has been funded in part by the U.S. Environmental Protection Agency. The authors declare they have no actual or potential competing financial interests. This study has been subjected to review by the Center for Computational Toxicology and Exposure and approved for publication. Approval does not signify that the contents reflect the views of the Agency, nor does mention of trade names or commercial products constitute endorsement or recommendation for use.
Figures
References
-
- Altenburger R, Backhaus T, Boedeker W et al (2000) Predictability of the toxicity of multiple chemical mixtures to Vibrio Fischeri: mixtures composed of similarly acting chemicals. Environ Tox Chem 19:2341–2347. 10.1002/etc.5620190926
-
- Atlas E, Bojanowski K, Mehmi I et al (2003) A deletion mutant of heregulin increases the sensitivity of breast cancer cells to chemotherapy without promoting tumorigenicity. Oncogene 22:3441–3451. 10.1038/sj.onc.1206410 - PubMed
-
- Beal MA, Coughlan MC, Nunnikhoven A et al (2024) High-throughput transcriptomics toxicity assessment of eleven data-poor bisphenol A alternatives. Environ Pollut. 10.1016/j.envpol.2024.124827 - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

