Early ctDNA and Survival in Metastatic Colorectal Cancer Treated With Immune Checkpoint Inhibitors: A Secondary Analysis of the SAMCO-PRODIGE 54 Randomized Clinical Trial
- PMID: 40531517
- PMCID: PMC12177728
- DOI: 10.1001/jamaoncol.2025.1646
Early ctDNA and Survival in Metastatic Colorectal Cancer Treated With Immune Checkpoint Inhibitors: A Secondary Analysis of the SAMCO-PRODIGE 54 Randomized Clinical Trial
Abstract
Importance: Immune checkpoint inhibitors (ICIs) have dramatically transformed the therapeutic landscape of deficient mismatch repair/microsatellite unstable-high (dMMR/MSI-H) metastatic colorectal cancer (mCRC); however, ICI use is challenged by primary resistance and timing of discontinuation. Whether circulating tumor DNA (ctDNA) may be predictive of progression-free survival (PFS) and overall survival (OS) in this treatment context remains unknown.
Objective: To assess the prognostic and predictive role of ctDNA, detected by tumor-specific methylation markers, in patients with dMMR/MSI-H mCRC treated with ICIs.
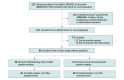
Design, setting, and participants: This prespecified secondary analysis of the SAMCO-PRODIGE 54 randomized clinical trial evaluated ctDNA in patients with dMMR/MSI-H mCRC treated with avelumab or standard chemotherapy, with or without a targeted agent in the second-line setting, to assess its prognostic role. Plasma samples were obtained prospectively for ctDNA analysis, and digital droplet polymerase chain reaction amplification of bisulfite-converted cell-free DNA (cfDNA) for WIF1 and NPY genes was used to quantify ctDNA levels. These samples were collected from April 2018 to April 2021 at 49 sites in France at baseline (V1) and 1-month posttreatment initiation (V2) during. Data analyses were performed from October 1 to November 1, 2024.
Intervention: Avelumab or standard chemotherapy with or without targeted agents.
Main outcomes and measures: PFS and OS according to baseline ctDNA positivity or concentration, and early ctDNA variation (ΔctDNA = [V1-V2] ÷ V1).
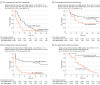
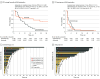
Results: The predictive analysis included 99 patients (mean [SD] age, 66 [13] years; 51 female [51.5%]) with plasma samples available for ctDNA assessment at V1, of which 74 had samples available also at V2 for Change in ctDNA assessment. In the 99 patients with available V1 plasma samples, baseline ctDNA positivity or concentration were not associated with clinical outcomes. Change in ctDNA (cutoff at median value) was significantly associated with both PFS (hazard ratio [HR], 2.98; 95% CI, 1.77-5.01; P < .001) and OS (HR, 3.61; 95% CI, 1.81-7.17; P < .001). This association was evident in patients treated with avelumab (PFS HR, 4.22; 95% CI, 1.77-10.1; P = .001; OS HR, 17.40; 95% CI, 3.82-79.70; P < .001) than in those receiving chemotherapy (PFS HR, 2.09; 95% CI, 1.03-4.21; P = .04; OS HR, 1.51; 95% CI, 0.61-3.72; P = .38). Avelumab (vs chemotherapy) improved PFS in favorable ctDNA responders (HR, 0.33; 95% CI, 0.14-0.77; log-rank P = .008) but not in poor responders (HR, 1.32; 95% CI, 0.67-2.62; log-rank P = .42) Combined ctDNA response and RECIST, version 1.1, assessment accurately predicted long-term OS. In the multivariable analysis, lack of ctDNA response was associated with an increased risk of disease progression and death in the avelumab group (HR, 7.27; 95% CI, 2.23-23.7; P = .001) but not in the chemotherapy group (HR, 1.61; 95% CI, 0.66-3.93; P = .30).
Conclusions: The findings of this secondary analysis of an RCT found that change in ctDNA at 1-month posttreatment can predict long-term outcomes in patients with dMMR/MSI-H mCRC treated with ICIs.
Trial registration: ClinicalTrials.gov Identifier: NCT03186326.
Conflict of interest statement
Figures
References
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

