Plexin/Semaphorin antagonism orchestrates collective cell migration and organ sculpting by regulating epithelial-mesenchymal balance
- PMID: 40532000
- PMCID: PMC12175886
- DOI: 10.1126/sciadv.adu3741
Plexin/Semaphorin antagonism orchestrates collective cell migration and organ sculpting by regulating epithelial-mesenchymal balance
Abstract
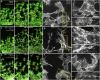
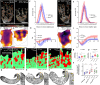
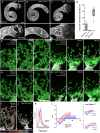
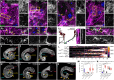
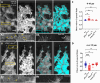
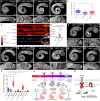
Cell behavior emerges from the intracellular distribution of properties such as protrusion, contractility, and adhesion. Thus, characteristic emergent rules of collective migration can arise from cell-cell contacts locally tweaking architecture, orchestrating self-regulation during development, wound healing, and cancer progression. The Drosophila testis-nascent-myotube system allows dissection of contact-dependent migration in vivo at high resolution. Here, we describe a role for the axon guidance factor Plexin A in collective cell migration: maintaining cell-cell interfaces at a precise point on the mesenchymal-to-epithelial continuum. This is crucial for testis myotubes to migrate as a continuous sheet, allowing normal sculpting-morphogenesis. Cells must maintain filopodial N-cadherin-based junctions and remain ECM-tethered near cell-cell contacts to spread while collectively moving. Our data further suggest Semaphorin 1b is a Plexin A antagonist, fine-tuning activation. This reveals a contact-dependent mechanism to maintain sheet integrity during migration, driving organ morphogenesis. This is relevant for mesenchymal organ sculpting in other migratory contexts such as angiogenesis.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

