Experimental evolution of a pathogen confronted with innate immune memory increases variation in virulence
- PMID: 40532126
- PMCID: PMC12176410
- DOI: 10.1371/journal.ppat.1012839
Experimental evolution of a pathogen confronted with innate immune memory increases variation in virulence
Abstract
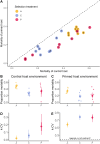
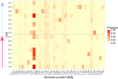
Understanding the drivers and mechanisms of virulence evolution is still a major goal of evolutionary biologists and epidemiologists. Theory predicts that the way virulence evolves depends on the balance between the benefits and costs it provides to pathogen fitness. Additionally, host responses to infections, such as resistance or tolerance, play a critical role in shaping virulence evolution. But, while the evolution of pathogens has been traditionally studied under the selection pressure of host adaptive immunity, less is known about their evolution when confronted to simpler and less effective forms of immunity such as immune priming. In this study, we used a well-established insect model for immune priming - red flour beetles and their bacterial pathogen Bacillus thuringiensis tenebrionis - to test how this form of innate immune memory drives the pathogen evolution. Through controlled experimental evolution of the pathogen in primed versus non-primed hosts, we found no change in average virulence after eight selection cycles in primed host. Nonetheless, we observed a notable rise in the variability of virulence, defined as the ability to kill hosts, among independent pathogen lines that evolved in primed hosts, and the bacteria were unable to develop resistance to host priming. Whole genome sequencing revealed increased activity in the bacterial mobilome (prophages and plasmids). Expression of the Cry toxin - a well-known virulence factor - was linked to evolved differences in copy number variation of the cry-carrying plasmid, though this did not correlate directly with virulence. These findings highlight that innate immune memory can drive variability in pathogen traits, which may favor adaptation to variable environments. This underscores the need to consider pathogen evolution in response to innate immune memory when applying these mechanisms in medicine, aquaculture, pest control, and insect mass production.
Copyright: © 2025 Korša et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





Similar articles
-
Immune priming in the insect gut: a dynamic response revealed by ultrastructural and transcriptomic changes.BMC Biol. 2025 Jul 28;23(1):227. doi: 10.1186/s12915-025-02334-4. BMC Biol. 2025. PMID: 40722160 Free PMC article.
-
Host and antibiotic jointly select for greater virulence in Staphylococcus aureus.bioRxiv [Preprint]. 2025 Apr 8:2024.08.31.610628. doi: 10.1101/2024.08.31.610628. bioRxiv. 2025. PMID: 39257827 Free PMC article. Preprint.
-
Links between Innate and Adaptive Immunity Can Favor Evolutionary Persistence of Immunopathology.Integr Comp Biol. 2024 Sep 27;64(3):841-852. doi: 10.1093/icb/icae105. Integr Comp Biol. 2024. PMID: 39030049 Free PMC article.
-
Trained Innate Immunity.Adv Exp Med Biol. 2025;1476:275-296. doi: 10.1007/978-3-031-85340-1_11. Adv Exp Med Biol. 2025. PMID: 40622547 Review.
-
Role of PE family of proteins in mycobacterial virulence: Potential on anti-TB vaccine and drug design.Int Rev Immunol. 2025;44(4):213-228. doi: 10.1080/08830185.2025.2455161. Epub 2025 Jan 31. Int Rev Immunol. 2025. PMID: 39889764 Review.
References
-
- Mackinnon MJ, Gandon S, Read AF. Virulence evolution in response to vaccination: The case of malaria. Vaccine. 2008;26(Suppl 3(48-5)): C42–C52. doi: . doi:10.1016/j.vaccine.2008.04.012. - DOI - PMC - PubMed
-
- Schmid-Hempel P. Evolutionary Parasitology: The Integrated Study of Infections, Immunology, Ecology, and Genetics [Internet]. Oxford University Press; 2013. [cited 2023 Jul 18]. Available from: doi: 10.1093/acprof:oso/9780199229482.001.0001 - DOI
MeSH terms
LinkOut - more resources
Full Text Sources

