PUMA reduces FASN ubiquitination to promote lipid accumulation and tumor progression in human clear cell renal cell carcinoma
- PMID: 40533456
- PMCID: PMC12177072
- DOI: 10.1038/s41419-025-07782-y
PUMA reduces FASN ubiquitination to promote lipid accumulation and tumor progression in human clear cell renal cell carcinoma
Abstract
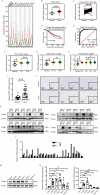
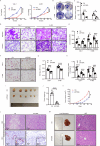
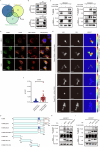
While the p53 upregulated modulator of apoptosis (PUMA) is traditionally recognized for promoting cell apoptosis and enhancing chemotherapy efficacy in various cancers, its role in clear cell renal cell carcinoma (ccRCC) remains unclear due to ccRCC's chemotherapy resistance. In this study, we discover a novel oncogenic role for PUMA in ccRCC, diverging from its known apoptotic function, through assessments of public datasets, clinical tissue samples, and cell line experiments. Abnormally high expression of PUMA positively correlates with clinical stages and poor prognosis. Notably, PUMA's role in ccRCC appears to be independent of apoptosis. Instead, it facilitates tumor progression and lipid accumulation through mechanisms involving the key metabolic regulator, fatty acid synthase (FASN). Specifically, the N44-102 amino acid sequence of PUMA, distinct from the previously studied BH3 domain, is crucial for its interaction with FASN. As a mechanism, PUMA stabilizes FASN by binding to ubiquitin-specific protease 15 (USP15), reducing FASN ubiquitination and degradation, thereby forming the PUMA-USP15-FASN axis. These findings challenge the established view of PUMA's role in cancer biology. Furthermore, PUMA knockdown significantly inhibits tumor growth and enhances the sensitivity of ccRCC tumors to metabolic inhibition. These results position PUMA as a novel metabolic regulator and a potential therapeutic target in ccRCC. The combined inhibition of PUMA and FASN further supports the therapeutic potential of targeting this metabolic axis.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests. Ethics approval and Consent to participate: (a) All methods in this study were performed in accordance with the relevant guidelines and regulations. (b) For animal experiments, approval was obtained from the Institutional Animal Care and Use Committee (IACUC) of Huazhong University of Science and Technology with approval number 3617. For human studies, approval was obtained from the Institutional Ethics Committee (IEC) of Tongji Medical College, Huazhong University of Science and Technology, Union Hospital with the reference number 0847. (c) Informed consent was obtained from all participants involved in the human studies. (d) No identifiable images from human research participants were included in this study.
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

