Reversible proliferative arrest induced by rapid depletion of RNase MRP
- PMID: 40533478
- PMCID: PMC12177063
- DOI: 10.1038/s41467-025-60471-4
Reversible proliferative arrest induced by rapid depletion of RNase MRP
Abstract
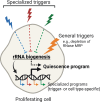
Cellular quiescence is a state of reversible proliferative arrest that plays essential roles in development, resistance to stress, aging, and longevity of organisms. Here we report that rapid depletion of RNase MRP, a deeply conserved RNA-based enzyme required for rRNA biosynthesis, induces a long-term yet reversible proliferative arrest in human cells. Severely compromised biogenesis of rRNAs along with acute transcriptional reprogramming precede a gradual decline of the critical cellular functions. Unexpectedly, many arresting cells show increased levels of histone mRNAs, which accumulate locally in the cytoplasm, and S-phase DNA amount. The ensuing proliferative arrest is entered from multiple stages of the cell cycle and can last for several weeks with uncompromised cell viability. Strikingly, restoring expression of RNase MRP leads to a complete reversal of the arrested state with resumed cell proliferation at the speed of control cells. We suggest that targeting rRNA biogenesis may provide a general strategy for rapid induction of a reversible proliferative arrest, with implications for understanding and manipulating cellular quiescence.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: J.M., W.F.M., and S.C. are inventors on a patent application filed by The Regents of the University of California (U.S. Provisional Patent Application No. 63/581,832) describing the methods of inducing reversible proliferative arrest presented in the manuscript. J.M. and S.C. are the founders of StopTime Inc., a startup focused on identifying and developing methods and compounds for the induction of cell dormancy and hypometabolism. All authors declare no other competing interests.
Figures



Similar articles
-
Composition and RNA binding specificity of metazoan RNase MRP.Nucleic Acids Res. 2025 Aug 27;53(16):gkaf829. doi: 10.1093/nar/gkaf829. Nucleic Acids Res. 2025. PMID: 40867056 Free PMC article.
-
Role of the SAF-A/HNRNPU SAP domain in X chromosome inactivation, nuclear dynamics, transcription, splicing, and cell proliferation.PLoS Genet. 2025 Jun 10;21(6):e1011719. doi: 10.1371/journal.pgen.1011719. eCollection 2025 Jun. PLoS Genet. 2025. PMID: 40493679 Free PMC article.
-
Prediction, screening and characterization of novel bioactive tetrapeptide matrikines for skin rejuvenation.Br J Dermatol. 2024 Jun 20;191(1):92-106. doi: 10.1093/bjd/ljae061. Br J Dermatol. 2024. PMID: 38375775
-
Assessing the comparative effects of interventions in COPD: a tutorial on network meta-analysis for clinicians.Respir Res. 2024 Dec 21;25(1):438. doi: 10.1186/s12931-024-03056-x. Respir Res. 2024. PMID: 39709425 Free PMC article. Review.
-
Pharmacological and electronic cigarette interventions for smoking cessation in adults: component network meta-analyses.Cochrane Database Syst Rev. 2023 Sep 12;9(9):CD015226. doi: 10.1002/14651858.CD015226.pub2. Cochrane Database Syst Rev. 2023. PMID: 37696529 Free PMC article.
Cited by
-
Composition and RNA binding specificity of metazoan RNase MRP.Nucleic Acids Res. 2025 Aug 27;53(16):gkaf829. doi: 10.1093/nar/gkaf829. Nucleic Acids Res. 2025. PMID: 40867056 Free PMC article.
References
-
- Dworkin, J. & Harwood, C. S. Metabolic reprogramming and longevity in quiescence. Annu. Rev. Microbiol.76, 91–111 (2022). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

