USP37 prevents premature disassembly of stressed replisomes by TRAIP
- PMID: 40533495
- PMCID: PMC12177040
- DOI: 10.1038/s41467-025-60139-z
USP37 prevents premature disassembly of stressed replisomes by TRAIP
Abstract
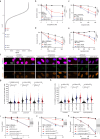
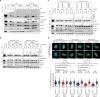
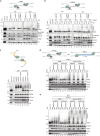
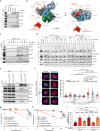
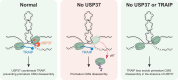
The eukaryotic replisome, which consists of the CDC45-MCM2-7-GINS (CMG) helicase, replicative polymerases, and several accessory factors, sometimes encounters proteinaceous obstacles that threaten genome integrity. These obstacles are targeted for removal or proteolysis by the E3 ubiquitin ligase TRAIP, which associates with the replisome. However, TRAIP must be carefully regulated to avoid inappropriate ubiquitylation and disassembly of the replisome. Here, we demonstrate that human cells lacking the de-ubiquitylating enzyme USP37 are hypersensitive to topoisomerase poisons and other replication stress-inducing agents. Furthermore, TRAIP loss rescues the hypersensitivity of USP37 knockout cells to topoisomerase inhibitors. In Xenopus egg extracts depleted of USP37, TRAIP promotes premature CMG ubiquitylation and disassembly when converging replisomes stall. Finally, guided by AlphaFold-Multimer, we discovered that binding to CDC45 mediates USP37's response to topological stress. We propose that USP37 protects genome stability by preventing TRAIP-dependent CMG unloading when replication stress impedes timely termination.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: J.C.W. is a co-founder of MOMA Therapeutics, in which he has a financial interest. S.P.J. is Chief Research Officer (part-time) at Insmed Innovation UK, Ltd. and founding partner of Ahren Innovation Capital LLP. He is a board member of Mission Therapeutics Ltd. and is a consultant and shareholder of Inflex Ltd. The remaining authors declare no competing interests.
Figures

Update of
-
USP37 prevents premature disassembly of stressed replisomes by TRAIP.bioRxiv [Preprint]. 2024 Sep 4:2024.09.03.611025. doi: 10.1101/2024.09.03.611025. bioRxiv. 2024. Update in: Nat Commun. 2025 Jun 18;16(1):5333. doi: 10.1038/s41467-025-60139-z. PMID: 39282314 Free PMC article. Updated. Preprint.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

