A molecular signature predicts hematologic evolution in polycythemia vera patients
- PMID: 40533498
- PMCID: PMC12310540
- DOI: 10.1038/s41375-025-02660-0
A molecular signature predicts hematologic evolution in polycythemia vera patients
Abstract
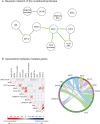
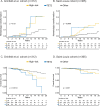
Genetic analyses have been included in scoring systems to improve the prognostic stratification of hematologic malignancies. Until now, molecular risk scores have not been included into the practical management of patients with polycythemia vera (PV). In this work, we studied 439 PV patients recruited from 15 French centers and described their mutational landscape using high-throughput sequencing. We detected an additional mutation in 53.3% of patients, 22.7% of them having 2 or more mutations. A Bayesian approach identified preferential associations between mutations. Based on these associations, we identified high molecular risk abnormalities in PV (PV-HMR), consisting in mutations in SRSF2, IDH1/2, EZH2 or NFE2 genes, copy number variations (CNV) and carrying 2 or more non-driver mutations. These PV-HMR were associated with decreased overall survival (OS) and/or transformation-free survival (TFS). Notably, ASXL1 mutations were not associated with a pejorative impact on OS or TFS when isolated. Based on these results, we developed a genomic 3-tier classification that efficiently predicted OS and more importantly TFS independently of age, sex, history of thrombosis and leukocyte and platelet counts. This model outperformed the IWG-PV and MIPSS-PV scoring systems in predicting the hematologic evolution of PV patients, which was confirmed in 2 external cohorts.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing financial interests. Ethics approval and patient consent: This work is also a deliverable of the FIMBANK network, which was founded by the French ‘Institut National du Cancer’ (INCa BCB 2013 & 2020).
Figures



References
-
- Vainchenker W, Kralovics R. Genetic basis and molecular pathophysiology of classical myeloproliferative neoplasms. Blood. 2017;129:667–79. - PubMed
-
- Hultcrantz M, Wilkes SR, Kristinsson SY, Andersson TM-L, Derolf ÅR, Eloranta S, et al. Risk and cause of death in patients diagnosed with myeloproliferative neoplasms in Sweden between 1973 and 2005: a population-based study. J Clin Oncol. 2015;33:2288–95. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

