Incorporating genetic data improves target trial emulations and informs the use of polygenic scores in randomized controlled trial design
- PMID: 40533517
- PMCID: PMC12283355
- DOI: 10.1038/s41588-025-02229-8
Incorporating genetic data improves target trial emulations and informs the use of polygenic scores in randomized controlled trial design
Erratum in
-
Author Correction: Incorporating genetic data improves target trial emulations and informs the use of polygenic scores in randomized controlled trial design.Nat Genet. 2025 Aug;57(8):2074. doi: 10.1038/s41588-025-02275-2. Nat Genet. 2025. PMID: 40579539 Free PMC article. No abstract available.
Abstract
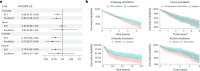
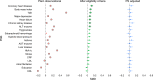
Randomized controlled trials (RCTs) remain the gold standard for evaluating medical interventions, yet ethical, practical and financial constraints often necessitate reliance on observational data and trial emulations. This study explores how integrating genetic data can enhance both emulated and traditional trial designs. Using FinnGen (n = 425,483), we emulated four major cardiometabolic RCTs and showed how reduced differences in polygenic scores (PGS) between trial arms track improvement in study design. Simulation studies reveal that PGS alone cannot fully adjust for unmeasured confounding. Instead, Mendelian randomization analyses can be used to detect likely confounders. Finally, trial emulations provide a platform to assess and refine PGS implementation for genetic enrichment strategies. By comparing associations of PGS with trial outcomes in the general population and emulated trial cohorts, we highlight the need to validate prognostic enrichment approaches in trial-relevant populations. These results highlight the growing potential of incorporating genetic information to optimize clinical trial design.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: A.G. is the founder of Real World Genetics OY. P.N. reports research grants from Allelica, Amgen, Apple, Boston Scientific, Genentech/Roche and Novartis; personal fees from Allelica, Apple, AstraZeneca, Blackstone Life Sciences, Creative Education Concepts, CRISPR Therapeutics, Eli Lilly & Co, Foresite Labs, Genentech/Roche, GV, HeartFlow, Magnet Biomedicine, Merck and Novartis; scientific advisory board membership of Esperion Therapeutics, Preciseli, TenSixteen Bio and Tourmaline Bio; scientific cofounder of TenSixteen Bio; equity in MyOme, Preciseli and TenSixteen Bio; and spousal employment at Vertex Pharmaceuticals (unrelated to the present work). A.P. is an employee of Alphabet and has previously received research support from Microsoft, Alphabet, Intel, IBM and Bayer. E.P. was supported by research grants from the National Institute of Diabetes and Digestive and Kidney Diseases (R01DK138036), the Patient Centered Outcomes Research Institute (DB-2020C2-20326) and the Food and Drug Administration (5U01FD007213), not related to the topic of this work. She is the principal investigator of a research grant to the Brigham and Women’s Hospital from Boehringer Ingelheim, not related to the topic of this work. She receives royalties from UpToDate. The other authors declare no competing interests.
Figures




Similar articles
-
Healthcare outcomes assessed with observational study designs compared with those assessed in randomized trials.Cochrane Database Syst Rev. 2014 Apr 29;2014(4):MR000034. doi: 10.1002/14651858.MR000034.pub2. Cochrane Database Syst Rev. 2014. Update in: Cochrane Database Syst Rev. 2024 Jan 4;1:MR000034. doi: 10.1002/14651858.MR000034.pub3. PMID: 24782322 Free PMC article. Updated.
-
Eliciting adverse effects data from participants in clinical trials.Cochrane Database Syst Rev. 2018 Jan 16;1(1):MR000039. doi: 10.1002/14651858.MR000039.pub2. Cochrane Database Syst Rev. 2018. PMID: 29372930 Free PMC article.
-
Audit and feedback: effects on professional practice.Cochrane Database Syst Rev. 2025 Mar 25;3(3):CD000259. doi: 10.1002/14651858.CD000259.pub4. Cochrane Database Syst Rev. 2025. PMID: 40130784
-
Conceptual framework and systematic review of the effects of participants' and professionals' preferences in randomised controlled trials.Health Technol Assess. 2005 Sep;9(35):1-186, iii-iv. doi: 10.3310/hta9350. Health Technol Assess. 2005. PMID: 16153352
-
Antidepressant treatment for postnatal depression.Cochrane Database Syst Rev. 2014 Sep 11;2014(9):CD002018. doi: 10.1002/14651858.CD002018.pub2. Cochrane Database Syst Rev. 2014. PMID: 25211400 Free PMC article.
References
-
- Feinstein Alvan, R. & Horwitz Ralph, I. Double standards, scientific methods, and epidemiologic research. N. Engl. J. Med.307, 1611–1617 (1982). - PubMed
-
- Real-World Evidence (U.S. Food and Drug Administration, 2024); www.fda.gov/science-research/science-and-research-special-topics/real-wo...
-
- Didelez, V., Haug, U. & Garcia-Albeniz, X. Re: are target trial emulations the gold standard for observational studies? Epidemiology35, e3 (2024). - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

