Insights from human NF-κB knockouts
- PMID: 40533538
- PMCID: PMC12287409
- DOI: 10.1038/s44319-025-00500-x
Insights from human NF-κB knockouts
Abstract
The well-studied NF-κB signaling system is a key mediator of the inflammatory response. Large-scale sequencing studies in humans now allow initial insights into non-essential human genes in which both alleles carry mutations that prevent protein expression or function. Here, we compiled the non-essential genes identified in various sequencing studies and analyzed the occurrence of knockouts in the human NF-κB signaling system. This revealed a lower knockout frequency in the NF-κB system compared to the entire genome. Since drugs inhibiting NF-κB pathway components were unsuccessful in clinical trials so far, the naturally occurring knockouts of NF-κB and its upstream regulators could provide new candidates for therapeutic intervention. To investigate the potential functional importance of posttranslational modifications (PTMs) occurring on NF-κB components, we analyzed not only their evolutionary conservation but also, as a second criterion, their genetic constraint in the sequenced individuals. This approach revealed the absence of missense mutations at key modification sites involved in NF-κB activation and identified additional candidate sites for future studies.
Keywords: Gene Essentiality; Knockout; Loss-of-function; NF-κB; Signaling.
© 2025. The Author(s).
Conflict of interest statement
Disclosure and competing interests statement. The authors declare no competing interests.
Figures
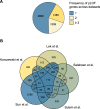
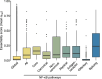
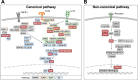


References
-
- Adeeb F, Dorris ER, Morgan NE, Lawless D, Maqsood A, Ng WL, Killeen O, Cummins EP, Taylor CT, Savic S et al (2021) A novel RELA truncating mutation in a familial Behcet’s disease-like mucocutaneous ulcerative condition. Arthritis Rheumatol 73:490–497 - PubMed
-
- Ayadi A, Birling MC, Bottomley J, Bussell J, Fuchs H, Fray M, Gailus-Durner V, Greenaway S, Houghton R, Karp N et al (2012) Mouse large-scale phenotyping initiatives: overview of the European Mouse Disease Clinic (EUMODIC) and of the Wellcome Trust Sanger Institute Mouse Genetics Project. Mamm Genome 23:600–610 - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

