Single-cell transcriptomic and chromatin dynamics of the human brain in PTSD
- PMID: 40533550
- PMCID: PMC12267058
- DOI: 10.1038/s41586-025-09083-y
Single-cell transcriptomic and chromatin dynamics of the human brain in PTSD
Abstract
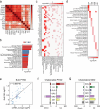
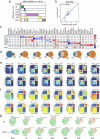
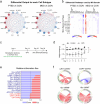
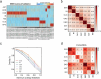
Post-traumatic stress disorder (PTSD) is a polygenic disorder occurring after extreme trauma exposure. Recent studies have begun to detail the molecular biology of PTSD. However, given the array of PTSD-perturbed molecular pathways identified so far1, it is implausible that a single cell type is responsible. Here we profile the molecular responses in over two million nuclei from the dorsolateral prefrontal cortex of 111 human brains, collected post-mortem from individuals with and without PTSD and major depressive disorder. We identify neuronal and non-neuronal cell-type clusters, gene expression changes and transcriptional regulators, and map the epigenomic regulome of PTSD in a cell-type-specific manner. Our analysis revealed PTSD-associated gene alterations in inhibitory neurons, endothelial cells and microglia and uncovered genes and pathways associated with glucocorticoid signalling, GABAergic transmission and neuroinflammation. We further validated these findings using cell-type-specific spatial transcriptomics, confirming disruption of key genes such as SST and FKBP5. By integrating genetic, transcriptomic and epigenetic data, we uncovered the regulatory mechanisms of credible variants that disrupt PTSD genes, including ELFN1, MAD1L1 and KCNIP4, in a cell-type-specific context. Together, these findings provide a comprehensive characterization of the cell-specific molecular regulatory mechanisms that underlie the persisting effects of traumatic stress response on the human prefrontal cortex.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: J.H.K. has consulting agreements (less than US$10,000 per year) with Aptinyx, Biogen, Idec, MA, Bionomics, Boehringer Ingelheim International, Epiodyne, EpiVario, Janssen Research and Development, Jazz Pharmaceuticals, Otsuka America Pharmaceutical, Spring Care, Sunovion Pharmaceuticals; is the co-founder for Freedom Biosciences; serves on the scientific advisory boards of Biohaven Pharmaceuticals, BioXcel Therapeutics (Clinical Advisory Board), Cerevel Therapeutics, Delix Therapeutics, Eisai, EpiVario, Jazz Pharmaceuticals, Neumora Therapeutics, Neurocrine Biosciences, Novartis Pharmaceuticals Corporation, PsychoGenics, Takeda Pharmaceuticals, Tempero Bio, Terran Biosciences; has stock options with Biohaven Pharmaceuticals Medical Sciences, Cartego Therapeutics, Damona Pharmaceuticals, Delix Therapeutics, EpiVario, Neumora Therapeutics, Rest Therapeutics, Tempero Bio, Terran Biosciences, Tetricus; and is an editor of Biological Psychiatry with income greater than US$10,000. The other authors declare no competing interests.
Figures










References
-
- Bromet, E., Sonnega, A. & Kessler, R. C. Risk factors for DSM-III-R posttraumatic stress disorder: findings from the National Comorbidity Survey. Am. J. Epidemiology147, 353–361 (1998). - PubMed
-
- Afifi, T. O., Asmundson, G. J. G., Taylor, S. & Jang, K. L. The role of genes and environment on trauma exposure and posttraumatic stress disorder symptoms: a review of twin studies. Clin. Psychol. Rev.30, 101–112 (2010). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

