R9AP is a common receptor for EBV infection in epithelial cells and B cells
- PMID: 40533557
- PMCID: PMC12328216
- DOI: 10.1038/s41586-025-09166-w
R9AP is a common receptor for EBV infection in epithelial cells and B cells
Abstract
Epstein-Barr virus (EBV) persistently infects more than 90% of the human population, causing infectious mononucleosis1, susceptibility to autoimmune diseases2 and multiple malignancies of epithelial or B cell-origin3. EBV infects epithelial cells and B cells through interaction between viral glycoproteins and different host receptors4, but it has remained unknown whether a common receptor mediates infection of its two major host cell targets. Here, we establish R9AP as a crucial EBV receptor for entry into epithelial and B cells. R9AP silencing or knockout, R9AP-derived peptide and R9AP monoclonal antibody each significantly inhibit, whereas R9AP overexpression promotes, EBV uptake into both cell types. R9AP binds directly to the EBV glycoprotein gH/gL complex to initiate gH/gL-gB-mediated membrane fusion. Notably, the interaction of R9AP with gH/gL is inhibited by the highly competitive gH/gL-neutralizing antibody AMMO1, which blocks EBV epithelial and B cell entry. Moreover, R9AP mediates viral and cellular membrane fusion in cooperation with EBV gp42-human leukocyte antigen class II or gH/gL-EPHA2 complexes in B cells or epithelial cells, respectively. We propose R9AP as the crucial common receptor of B cells and epithelial cells and a potential prophylactic and vaccine target for EBV.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: Patent applications related to the R9AP monoclonal antibody 5E9 have been submitted to the China National Intellectual Property Administration (patent application number 2024119195522; M.-S.Z., C.S. and C.X. are named inventors). The other authors declare no competing interests.
Figures










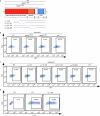
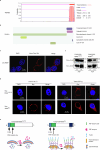
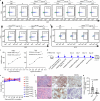
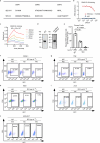

References
-
- Luzuriaga, K. & Sullivan, J. L. Infectious mononucleosis. N. Engl. J. Med.362, 1993–2000 (2010). - PubMed
-
- Bjornevik, K. et al. Longitudinal analysis reveals high prevalence of Epstein–Barr virus associated with multiple sclerosis. Science375, 296–301 (2022). - PubMed
-
- Zhong, L. Y. et al. Research landmarks on the 60th anniversary of Epstein–Barr virus. Sci. China Life Sci.68, 354–380 (2025). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

