Targeting de novo purine biosynthesis for tuberculosis treatment
- PMID: 40533558
- PMCID: PMC12328218
- DOI: 10.1038/s41586-025-09177-7
Targeting de novo purine biosynthesis for tuberculosis treatment
Abstract
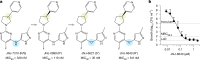
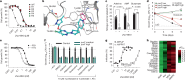
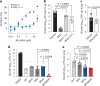
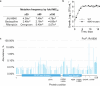
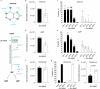
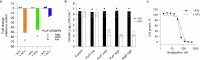
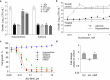
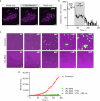
Tuberculosis remains the leading cause of death from an infectious disease1,2. Here we report the discovery of a first-in-class small-molecule inhibitor targeting PurF, the first enzyme in the mycobacterial de novo purine biosynthesis pathway. The lead candidate, JNJ-6640, exhibited nanomolar bactericidal activity in vitro. Comprehensive genetic and biochemical approaches confirmed that JNJ-6640 was highly selective for mycobacterial PurF. Single-cell-level microscopy demonstrated a downstream effect on DNA replication. We determined the physiologically relevant concentrations of nucleobases in human and mouse lung tissue, showing that these levels were insufficient to salvage PurF inhibition. Indeed, proof-of-concept studies using a long-acting injectable formulation demonstrated the in vivo efficacy of the compound. Finally, we show that inclusion of JNJ-6640 could have a crucial role in improving current treatment regimens for drug-resistant tuberculosis. Together, we demonstrate that JNJ-6640 is a promising chemical lead and that targeting de novo purine biosynthesis represents a novel strategy for tuberculosis drug development.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: D.A.L., A.L., J.S., P.F., J.W., S.D., S. Saylock, M.D.R., A.H., T.G., A.V., K.W.C., P.S., M.E., M.M., J.E., R.D.A., H.F., A.S.P., B.S., C.A.-P., P.J. and A.K. were or are all full-time employees of Janssen Pharmaceutica, a Johnson & Johnson company, and/or potential stockholders of Johnson & Johnson. J.S. and P.F. were or are employees of Charles River Laboratories, a contract research organization. A.R., C.R., N.C. and S. Sans were or are all full-time employees of Evotec. R.J.W., C.D., W.P., E.D., S.L., M.G., J.D., S.J.W., G.L.-M., N.D. and A.K. received funding from Janssen Pharmaceutica to perform contract research. The other authors declare no competing interests.
Figures




References
-
- TB alliance. Get the facts. TB Alliancehttps://www.tballiance.org/why-new-tb-drugs/global-pandemic (2023).
-
- World Health Organization. Global tuberculosis report 2023. WHOhttps://www.who.int/teams/global-tuberculosis-programme/tb-reports/globa... (2023).
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

