Allosteric modulation and biased signalling at free fatty acid receptor 2
- PMID: 40533560
- PMCID: PMC12570966
- DOI: 10.1038/s41586-025-09186-6
Allosteric modulation and biased signalling at free fatty acid receptor 2
Abstract
Free fatty acid receptor 2 (FFA2) is a G protein-coupled receptor (GPCR) that is a primary sensor for short-chain fatty acids produced by gut microbiota. Consequently, FFA2 is a promising drug target for immunometabolic disorders1-4. Here we report cryogenic electronic microscopy structures of FFA2 in complex with two G proteins and three distinct classes of positive allosteric modulators (PAMs), and describe noncanonical activation mechanisms that involve conserved structural features of class A GPCRs. Two PAMs disrupt the E/DRY activation microswitch5 and stabilize the conformation of intracellular loop 2 by binding to lipid-facing pockets near the cytoplasmic side of the receptor. By contrast, the third PAM promotes the separation of transmembrane helices 6 and 7 by interacting with transmembrane helix 6 at the receptor-lipid interface. Molecular dynamic simulations and mutagenesis experiments confirm these noncanonical activation mechanisms. Furthermore, we demonstrate the molecular basis for the Gi versus Gq bias, which is due to distinct conformations of intracellular loop 2 stabilized by different PAMs. These findings provide a framework for the design of tailored GPCR modulators, with implications that extend beyond FFA2 to the broader field of GPCR drug discovery.
© 2025. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
Competing interests: G.M. and T.U. are co-founders and directors of Caldan Therapeutics ( https://www.caldantherapeutics.com/ ). G.M. is a co-founder and director of KelticPharmaTherapeutics ( https://keltic-pharma.com/ ). Both companies have interests in the development of FFA4 activators. The other authors declare no competing interests.
Figures









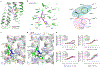



References
-
- Stoddart LA, Smith NJ & Milligan G International Union of Pharmacology. LXXI. Free fatty acid receptors FFA1, -2, and -3: pharmacology and pathophysiological functions. Pharmacol. Rev 60, 405–417 (2008). - PubMed
-
- Alvarez-Curto E & Milligan G Metabolism meets immunity: the role of free fatty acid receptors in the immune system. Biochem. Pharmacol 114, 3–13 (2016). - PubMed
-
- Kimura I, Ichimura A, Ohue-Kitano R & Igarashi M Free fatty acid receptors in health and disease. Physiol. Rev 100, 171–210 (2020). - PubMed
-
- McCarville JL, Chen GY, Cuevas VD, Troha K & Ayres JS Microbiota metabolites in health and disease. Annu. Rev. Immunol 38, 147–170 (2020). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

