Kupffer cell programming by maternal obesity triggers fatty liver disease
- PMID: 40533564
- PMCID: PMC12367551
- DOI: 10.1038/s41586-025-09190-w
Kupffer cell programming by maternal obesity triggers fatty liver disease
Abstract
Kupffer cells (KCs) are tissue-resident macrophages that colonize the liver early during embryogenesis1. Upon liver colonization, KCs rapidly acquire a tissue-specific transcriptional signature, mature alongside the developing liver and adapt to its functions1-3. Throughout development and adulthood, KCs perform distinct core functions that are essential for liver and organismal homeostasis, including supporting fetal erythropoiesis, postnatal erythrocyte recycling and liver metabolism4. However, whether perturbations of macrophage core functions during development contribute to or cause disease at postnatal stages is poorly understood. Here, we utilize a mouse model of maternal obesity to perturb KC functions during gestation. We show that offspring exposed to maternal obesity develop fatty liver disease, driven by aberrant developmental programming of KCs that persists into adulthood. Programmed KCs promote lipid uptake by hepatocytes through apolipoprotein secretion. KC depletion in neonate mice born to obese mothers, followed by replenishment with naive monocytes, rescues fatty liver disease. Furthermore, genetic ablation of the gene encoding hypoxia-inducible factor-α (HIF1α) in macrophages during gestation prevents the metabolic programming of KCs from oxidative phosphorylation to glycolysis, thereby averting the development of fatty liver disease. These results establish developmental perturbation of KC functions as a causal factor in fatty liver disease in adulthood and position fetal-derived macrophages as critical intergenerational messengers within the concept of developmental origins of health and diseases5.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures

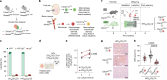
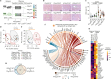

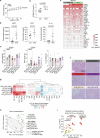

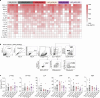





References
-
- Barker, D. J. P. The developmental origins of chronic adult disease. Acta Paediat. Suppl.10.1080/08035320410022730 (2004). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

