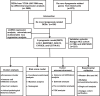
The impact of de novo lipogenesis on predicting survival and clinical therapy: an exploration based on a multigene prognostic model in hepatocellular carcinoma
- PMID: 40533802
- PMCID: PMC12178006
- DOI: 10.1186/s12967-025-06704-y
The impact of de novo lipogenesis on predicting survival and clinical therapy: an exploration based on a multigene prognostic model in hepatocellular carcinoma
Abstract
Background: Hepatocellular carcinoma (HCC) ranks among the most aggressive malignancies worldwide, with poor outcomes attributed to delayed diagnosis and therapeutic limitations. Emerging evidence suggests that de novo lipogenesis (DNL) plays a crucial role in HCC progression and its interaction with the immune microenvironment.
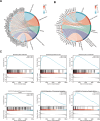
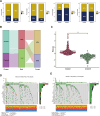
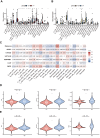
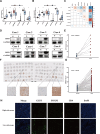
Methods: We systematically analyzed DNL-related gene expression profiles from TCGA, GEO, ICGC-LIRI datasets, and our Xiangya HCC cohort (n = 106) to construct a prognostic risk model. Through LASSO-Cox regression analysis, we identified six signature genes (G6PD, LCAT, SERPINE1, SOAT2, CYP2C9, and UGT1A10) that effectively stratified patients into distinct risk groups. We evaluated clinical characteristics, immune cell infiltration patterns, and differential therapeutic responses between high-risk and low-risk groups. Comprehensive validation included immunohistochemical analysis and Western blotting to assess expression levels of key model genes, along with multiplex immunofluorescence staining and single-cell RNA sequencing(scRNA-seq) to characterize immune microenvironmental differences between risk groups.
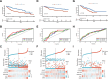
Results: We successfully established a robust six-gene prognostic signature (G6PD, LCAT, SERPINE1, SOAT2, CYP2C9, and UGT1A10) based on de novo lipogenesis pathways, which demonstrated excellent predictive performance (AUC: 0.78-0.82). The model revealed significant differences in immune infiltration patterns between risk groups, with the high-risk group exhibiting immunosuppressive characteristics characterized by increased Treg cell infiltration, while the low-risk group showed greater NK cell retention. Integrated scRNA-seq and our cohort validation further demonstrated that high-risk scores were associated with poorer response to immunotherapy but greater sensitivity to targeted therapies. These findings suggest that de novo lipogenesis-mediated immune evasion contributes to therapy resistance and worse prognosis in high-risk HCC patients, whereas low-risk HCC patients maintain an immunologically active microenvironment more amenable to immunotherapy.
Conclusions: This study provided a novel prognostic model for HCC, incorporating 6 representative DNLs. The model demonstrated the potential for predicting HCC prognosis and highlighted the involvement of immune cell infiltration and the association between risk scores and clinical therapy. Validation of model genes further supported the association between de novo lipogenesis and HCC development.
Keywords: Clinical therapy; De novo lipogenesis; Multigene prognostic model; RNA-seq; Single-cell RNA-seq.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: This study was approved by Ethics Committees of Xiangya Hospital and patient consent was obtained before the samples were taken. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures




Similar articles
-
Prognostic and Immunological Significance of the Molecular Subtypes and Risk Signatures Based on Cuproptosis in Hepatocellular Carcinoma.Mediators Inflamm. 2023 Apr 20;2023:3951940. doi: 10.1155/2023/3951940. eCollection 2023. Mediators Inflamm. 2023. PMID: 37124062 Free PMC article.
-
Identification of anoikis-related subtypes and a risk score prognosis model, the association with TME landscapes and therapeutic responses in hepatocellular carcinoma.Front Immunol. 2025 Jun 17;16:1602831. doi: 10.3389/fimmu.2025.1602831. eCollection 2025. Front Immunol. 2025. PMID: 40599775 Free PMC article.
-
A Novel RNA Methylation-Related Prognostic Signature and its Tumor Microenvironment Characterization in Hepatocellular Carcinoma.Technol Cancer Res Treat. 2024 Jan-Dec;23:15330338241276895. doi: 10.1177/15330338241276895. Technol Cancer Res Treat. 2024. PMID: 39155614 Free PMC article.
-
Deciphering adenosine signaling in hepatocellular carcinoma: Pathways, prognostic models, and therapeutic implications.Clin Mol Hepatol. 2025 Jul;31(3):706-729. doi: 10.3350/cmh.2024.1068. Epub 2025 Feb 5. Clin Mol Hepatol. 2025. PMID: 39905839 Free PMC article. Review.
-
The Protective Role of miR-125b in Hepatocellular Carcinoma: Unraveling Tumor-Suppressive Mechanisms.Curr Mol Med. 2025;25(6):663-671. doi: 10.2174/0115665240304247240529074123. Curr Mol Med. 2025. PMID: 38859784 Review.
References
-
- Bray F, Laversanne M, Sung H, Ferlay J, Siegel RL, Soerjomataram I, Jemal A. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2024;74(3):229–63. - PubMed
-
- Singal AG, Kanwal F, Llovet JM. Global trends in hepatocellular carcinoma epidemiology: implications for screening, prevention and therapy. Nat Rev Clin Oncol. 2023;20(12):864–84. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

