NRF2 maintains redox balance via ME1 and NRF2 inhibitor synergizes with venetoclax in NPM1-mutated acute myeloid leukemia
- PMID: 40533864
- PMCID: PMC12177962
- DOI: 10.1186/s40170-025-00401-6
NRF2 maintains redox balance via ME1 and NRF2 inhibitor synergizes with venetoclax in NPM1-mutated acute myeloid leukemia
Abstract
Background: Acute myeloid leukemia (AML) with nucleophosmin 1 (NPM1) mutations represents a distinct subtype of leukemia. Emerging evidence suggests that regulation of redox metabolism contributes to tumorigenesis and reveals a metabolic vulnerability in anti-tumor therapies. However, the role of redox homeostasis between reactive oxygen species (ROS) and antioxidant systems plays in NPM1-mutated AML has not been fully elucidated.
Methods: First, ROS-related metabolic pathways in NPM1-mutated AML were analyzed using RNA-sequencing data. Intracellular and mitochondrial ROS levels in leukemia cells were detected using flow cytometry (FCM). The expression of nuclear factor (erythroid-derived 2)-like 2 (NRF2) was analyzed in public databases and further validated in AML primary blasts and cell lines by quantitative real-time PCR (qRT-PCR), western blotting, and immunofluorescence. Next, the mechanism underlying NRF2 expression was investigated through the RNA immunoprecipitation (RIP), methylated RNA immunoprecipitation (MeRIP) and rescue experiments. Additionally, the downstream target gene of NRF2 was identified by bioinformatics analysis and chromatin immunoprecipitation (ChIP) assays. Furthermore, RNA interference and the NRF2 inhibitor ML385 were applied to explore the role of NRF2 in leukemia. Finally, the anti-leukemic effects of ML385 alone or in combination with the B-cell lymphoma 2 (BCL-2) inhibitor venetoclax on AML cells were investigated using FCM analysis and western blotting, and further explored in cell line-derived xenograft (CDX) mouse models.
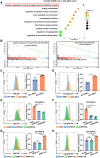
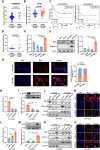
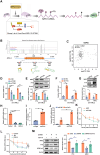
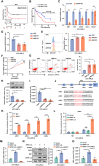
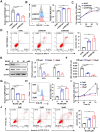
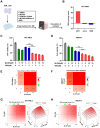
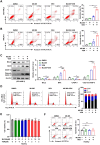
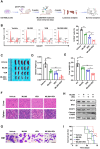
Results: In this study, we identified significant ROS accumulation in leukemia cells with NPM1 mutations. Meanwhile, elevated NRF2 expression and its nuclear localization were observed in NPM1-mutated AML cells. The high NRF2 expression levels were at least partially induced by fat mass and obesity-associated protein (FTO) via m6A modification. Functionally, NRF2 exerts its antioxidant effects by transcriptionally upregulating malic enzyme 1 (ME1) expression and enhancing its activity. Targeting NRF2/ME1 axis reduced NADPH/NADP+ ratio, increased ROS levels, impaired leukemia cell viability, and promoted apoptosis. More importantly, NRF2 inhibitor ML385 in combination with venetoclax showed synergistic anti-leukemic activity in vitro and in vivo.
Conclusion: Overall, our findings provide new insight into the therapeutic potential of targeting NRF2 and guide the development of innovative combination therapies in NPM1-mutated AML.
Keywords: Acute myeloid leukemia; ME1; NRF2; Nucleophosmin 1; Redox homeostasis; Venetoclax.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Ethical approval was given by the Medical Ethics Committee of Chongqing Medical University. All animal procedures to be employed in the project was approved by Institutional Animal Care and Use of Chongqing Medical University (Approval number: IACUC-CQMU-2025-0338). Competing interests: The authors declare no competing interests.
Figures
References
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

