Host traits and environmental variation shape gut microbiota diversity in wild threespine stickleback
- PMID: 40533873
- PMCID: PMC12177961
- DOI: 10.1186/s42523-025-00404-0
Host traits and environmental variation shape gut microbiota diversity in wild threespine stickleback
Abstract
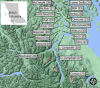
Background: Despite the growing recognition of the importance of gut microbiota in host ecology and evolution, our understanding of the relative contributions of host-associated and environmental factors shaping gut microbiota composition within and across wild populations remains limited. Here, we investigate how host morphology, sex, genetic divergence, and environmental characteristics influence the gut microbiota of threespine stickleback fish populations from 20 lakes on Vancouver Island, Canada.
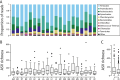
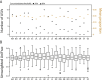
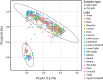
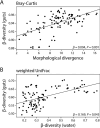
Results: Our findings reveal substantial variation in gut microbiota composition and diversity among populations, with host traits exerting a relatively stronger influence on bacterial alpha diversity than environmental characteristics. Previous studies have suggested a link between stickleback body shape and niche specialization, and our results indicate that aspects of host morphology may be associated with gut microbiota divergence among populations, though whether this is related to trophic ecology remains to be explored. Within and across populations, we only observed a weakly defined core microbiota and limited sharing of amplicon sequence variants (ASVs) among hosts, indicating that gut microbiota composition is individualized. Additionally, we detected sex-dependent differences in microbial diversity, opening avenues for future research into the mechanisms driving this variation.
Conclusions: In sum, our study emphasizes the need to consider both host-associated and environmental factors in shaping gut microbiota dynamics and highlights the complex interplay between host organisms, their associated microbial communities, and the environment in natural settings. Ultimately, these insights add to our understanding of the eco-evolutionary implications of host-microbiota interactions while underscoring the need for further investigation into the underlying mechanisms.
Keywords: Gasterosteus aculeatus; 16S rRNA sequencing; Animal microbiome; Gut microbiome.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Samples were collected in 2020, 2021, and 2022 under British Columbia Fish Collection permits NA20-602264, MRVI21-619908, and NA22-713085, respectively. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures
References
-
- Adams DC, Collyer ML, Kaliontzopoulou A, and Baken E. Geomorph: Software for geometric morphometric analyses.2021; R package version 3.3.2. https://cran.r-project.org/package=geomorph.
-
- Albert AY, Sawaya S, Vines TH, Knecht AK, Miller CT, Summers BR, Balabhadra S, Kingsley DM, Schluter D. The genetics of adaptive shape shift in stickleback: pleiotropy and effect size. Evolution. 2008;62:76–85. - PubMed
-
- Anderson MJ. A new method for non-parametric multivariate analysis of variance. Aust J Ecol. 2001;26:32–46.
Grants and funding
LinkOut - more resources
Full Text Sources
