Diamond Molecular Balance: Ultra-Wide Range Nanomechanical Mass Spectrometry from MDa to TDa
- PMID: 40534199
- PMCID: PMC12232386
- DOI: 10.1021/acs.nanolett.5c02032
Diamond Molecular Balance: Ultra-Wide Range Nanomechanical Mass Spectrometry from MDa to TDa
Abstract
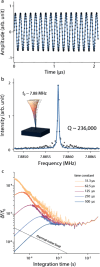
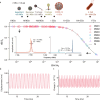
The significance of mass spectrometry lies in its unparalleled ability to accurately identify and quantify molecules in complex samples, providing invaluable insights into molecular structures and interactions. Here, we leverage diamond nanostructures as highly sensitive mass sensors by utilizing a self-excitation mechanism under an electron beam in a conventional scanning electron microscope (SEM). The diamond molecular balance (DMB) exhibits a practical mass resolution of 4.07 MDa, based on its notable mechanical quality factor and frequency stability, along with a broad dynamic range from MDa to TDa. This positions the DMB at the forefront of nanoelectromechanical system (NEMS)-based mass spectrometry operating at room temperature. Notably, the DMB demonstrated its ability to measure the mass of a single bacteriophage T4 by precisely locating the analyte on the device. These findings demonstrate the capability and potential of the DMB as a revolutionary tool for mass spectrometry at room temperature.
Keywords: Bacteriophage T4; Diamond; Mass Spectrometry; Multiplexing4; Nanoelectromechanical Systems.
Figures



Similar articles
-
Neuromuscular electrical stimulation (NMES) for patellofemoral pain syndrome.Cochrane Database Syst Rev. 2017 Dec 12;12(12):CD011289. doi: 10.1002/14651858.CD011289.pub2. Cochrane Database Syst Rev. 2017. PMID: 29231243 Free PMC article.
-
Comparison of Two Modern Survival Prediction Tools, SORG-MLA and METSSS, in Patients With Symptomatic Long-bone Metastases Who Underwent Local Treatment With Surgery Followed by Radiotherapy and With Radiotherapy Alone.Clin Orthop Relat Res. 2024 Dec 1;482(12):2193-2208. doi: 10.1097/CORR.0000000000003185. Epub 2024 Jul 23. Clin Orthop Relat Res. 2024. PMID: 39051924
-
Factors that impact on the use of mechanical ventilation weaning protocols in critically ill adults and children: a qualitative evidence-synthesis.Cochrane Database Syst Rev. 2016 Oct 4;10(10):CD011812. doi: 10.1002/14651858.CD011812.pub2. Cochrane Database Syst Rev. 2016. PMID: 27699783 Free PMC article.
-
A New Measure of Quantified Social Health Is Associated With Levels of Discomfort, Capability, and Mental and General Health Among Patients Seeking Musculoskeletal Specialty Care.Clin Orthop Relat Res. 2025 Apr 1;483(4):647-663. doi: 10.1097/CORR.0000000000003394. Epub 2025 Feb 5. Clin Orthop Relat Res. 2025. PMID: 39915110
-
Is Your Surgical Helmet System Compromising the Sterile Field? A Systematic Review of Contamination Risks and Preventive Measures in Total Joint Arthroplasty.Clin Orthop Relat Res. 2025 Jun 1;483(6):972-990. doi: 10.1097/CORR.0000000000003383. Epub 2025 Feb 5. Clin Orthop Relat Res. 2025. PMID: 39915114
References
LinkOut - more resources
Full Text Sources

