The genome sequence of the Violet Copper, Lycaena helle (Denis & Schiffermüller, 1775)
- PMID: 40534729
- PMCID: PMC12174908
- DOI: 10.12688/f1000research.156485.1
The genome sequence of the Violet Copper, Lycaena helle (Denis & Schiffermüller, 1775)
Abstract
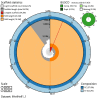
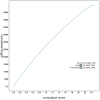
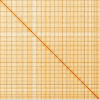
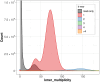
We present a genome assembly from an individual female Lycaena helle (the Violet Copper; Arthropoda; Insecta; Lepidoptera; Lycaenidae). The genome sequence is 547.31 megabases in span. The entirety of the genome sequence was assembled into 25 contiguous chromosomal pseudomolecules with no gaps, including the Z and W sex chromosomes. The mitochondrial genome has also been assembled and is 15.5 kilobases in length. Gene annotation of this assembly identified 20,122 protein coding genes.
Keywords: Lepidoptera; Lycaena helle; RNA-seq; Violet Copper; genome sequence.
Copyright: © 2025 Pladevall C et al.
Conflict of interest statement
No competing interests were disclosed.
Figures


Similar articles
-
The genome sequence of the Riband Wave, Idaea aversata (Linnaeus, 1758).Wellcome Open Res. 2023 Jan 31;8:45. doi: 10.12688/wellcomeopenres.18899.1. eCollection 2023. Wellcome Open Res. 2023. PMID: 37484483 Free PMC article.
-
The genome sequence of the Little Grey, Eudonia lacustrata (Panzer, 1804).Wellcome Open Res. 2023 Jul 11;8:302. doi: 10.12688/wellcomeopenres.19646.1. eCollection 2023. Wellcome Open Res. 2023. PMID: 39026606 Free PMC article.
-
The genome sequence of the Case-bearing Clothes moth, Tinea pellionella (Linnaeus, 1758).Wellcome Open Res. 2024 Nov 4;9:119. doi: 10.12688/wellcomeopenres.21015.2. eCollection 2024. Wellcome Open Res. 2024. PMID: 39494195 Free PMC article.
-
The genome sequence of the Small Emerald, Hemistola chrysoprasaria (Esper, 1795).Wellcome Open Res. 2023 Oct 12;8:441. doi: 10.12688/wellcomeopenres.19999.1. eCollection 2023. Wellcome Open Res. 2023. PMID: 39224879 Free PMC article.
-
The genome sequence of the Brindled Beauty, Lycia hirtaria (Clerck, 1759).Wellcome Open Res. 2023 Jul 12;8:303. doi: 10.12688/wellcomeopenres.19650.1. eCollection 2023. Wellcome Open Res. 2023. PMID: 39026607 Free PMC article.
References
-
- Habel JC, Schmitt T, Meyer M, et al. : Biogeography meets conservation: the genetic structure of the endangered lycaenid butterfly Lycaena helle (Denis & Schiffermüller, 1775). Biol. J. Linn. Soc. 2010 Sep;101(1):155–168. 10.1111/j.1095-8312.2010.01471.x - DOI
-
- Baguette M, Nève G: Adult movements between populations in the specialist butterfly Proclossiana eunomia (Lepidoptera, Nymphalidae). Ecol. Entomol. 1994 Feb;19(1):1–5. 10.1111/j.1365-2311.1994.tb00382.x - DOI
-
- Lesica P, Allendorf FW: When Are Peripheral Populations Valuable for Conservation? Conserv. Biol. 1995 Aug;9(4):753–760. 10.1046/j.1523-1739.1995.09040753.x - DOI
MeSH terms
Associated data
LinkOut - more resources
Full Text Sources

