MOLUNGN: a multi-omics graph neural network for biomarker discovery and accurate lung cancer classification
- PMID: 40534839
- PMCID: PMC12174459
- DOI: 10.3389/fgene.2025.1610284
MOLUNGN: a multi-omics graph neural network for biomarker discovery and accurate lung cancer classification
Abstract
Introduction: Lung cancer continues to pose significant global health burdens due to its high morbidity and mortality. This study aimed to systematically integrate biomedical datasets, particularly incorporating traditional Chinese medicine (TCM)-associated multi-omics data, employing advanced deep-learning methods enhanced by graph attention mechanisms. We sought to investigate molecular mechanisms underlying stage-wise lung cancer progression and identify pivotal stage-specific biomarkers to support precise cancer staging classification.
Methods: We developed a novel multi-omics integrative model, named the Multi-Omics Lung Cancer Graph Network (MOLUNGN), based on Graph Attention Networks (GAT). Clinical datasets of non-small cell lung cancer (NSCLC), including lung adenocarcinoma (LUAD) and lung squamous cell carcinoma (LUSC), were analyzed to create omics-specific feature matrices comprising mRNA expression, miRNA mutation profiles, and DNA methylation data. MOLUNGN incorporated omics-specific GAT modules (OSGAT) combined with a Multi-Omics View Correlation Discovery Network (MOVCDN), effectively capturing intra- and inter-omics correlations. This framework enabled comprehensive classification of clinical cases into precise cancer stages, alongside the extraction of stage-specific biomarkers.
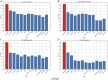
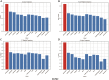
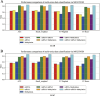
Results: Evaluations utilizing publicly available datasets confirmed MOLUNGN's superior performance over existing methodologies. On the LUAD dataset, MOLUNGN achieved accuracy (ACC) of 0.84, Recall_weighted of 0.84, F1_weighted of 0.83, and F1_macro of 0.82. On the LUSC dataset, the model further improved, achieving ACC of 0.86, Recall_weighted of 0.86, F1_weighted of 0.85, and F1_macro of 0.84. Notably, critical stage-specific biomarkers with significant biological relevance to lung cancer progression were identified, facilitating robust gene-disease associations.
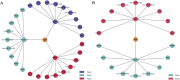
Discussion: Our findings underscore the efficacy of MOLUNGN as an integrative framework in accurately classifying lung cancer stages and uncovering essential biomarkers. These biomarkers provide deep insights into lung cancer progression mechanisms and represent promising targets for future clinical validation. Integrating these biomarkers into the TCM-target-disease network enriches the understanding of TCM therapeutic potentials, laying a robust foundation for future precision medicine applications.
Keywords: GAT; MOLUNGN; lung cancer; multi-omics data integration; stage prediction.
Copyright © 2025 Zhang, Bian, Zhang, Xie and Hu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
-
- Brierley J. D., Gospodarowicz M. K., Wittekind C. (2017). TNM classification of malignant tumours. John Wiley and Sons.
LinkOut - more resources
Full Text Sources

