KLF feedback loops in innate immunity
- PMID: 40534851
- PMCID: PMC12173922
- DOI: 10.3389/fimmu.2025.1606277
KLF feedback loops in innate immunity
Abstract
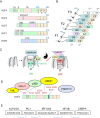
The Krüppel-like factor (KLF) family of zinc finger transcription factors regulate the expression of genes involved in a wide range of cellular processes, including cell proliferation and differentiation. In haematopoiesis, KLFs have essential roles in myeloid cell differentiation and function. KLF4 is a critical regulator of macrophage development and initiates pro- and anti-inflammatory signalling pathways in response to various stimuli. KLF2, KLF3 and KLF6 also play important roles in regulating these pathways. Here we review how KLFs cooperate and compete to either activate or repress target genes to influence initiation and resolution of inflammatory responses in macrophages. We also discuss how KLFs may be involved in the development of chronic inflammatory conditions.
Keywords: KLF3; KLF4; chronic inflammatory disease; feedback loops; inflammation; innate immunity; macrophage; transcriptional regulation.
Copyright © 2025 Salmon, Adams, Magor and Perkins.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Figures




Similar articles
-
Alternative splicing of KLF4 in myeloid cells: implications for cellular plasticity and trained immunity in cancer and inflammatory disease.Front Immunol. 2025 Jun 9;16:1585528. doi: 10.3389/fimmu.2025.1585528. eCollection 2025. Front Immunol. 2025. PMID: 40552304 Free PMC article. Review.
-
Krüppel-like factor 4 control of immune cell function.Front Immunol. 2025 Aug 4;16:1597210. doi: 10.3389/fimmu.2025.1597210. eCollection 2025. Front Immunol. 2025. PMID: 40831570 Free PMC article. Review.
-
The reprogramming factor KLF4 in normal and malignant blood cells.Front Immunol. 2025 Jun 16;16:1584181. doi: 10.3389/fimmu.2025.1584181. eCollection 2025. Front Immunol. 2025. PMID: 40589762 Free PMC article. Review.
-
Transcriptional profiling of lung macrophages following ozone exposure in mice identifies signaling pathways regulating immunometabolic activation.Toxicol Sci. 2024 Sep 1;201(1):103-117. doi: 10.1093/toxsci/kfae081. Toxicol Sci. 2024. PMID: 38897669 Free PMC article.
-
The kruppel-like factor (KLF) family, diseases, and physiological events.Gene. 2024 Feb 15;895:148027. doi: 10.1016/j.gene.2023.148027. Epub 2023 Nov 23. Gene. 2024. PMID: 38000704 Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

