T cell-mediated immune surveillance conferred by latent Epstein-Barr virus genes suppresses a broad spectrum of tumor formation through NKG2D-NKG2DL interactions
- PMID: 40534857
- PMCID: PMC12174148
- DOI: 10.3389/fimmu.2025.1597731
T cell-mediated immune surveillance conferred by latent Epstein-Barr virus genes suppresses a broad spectrum of tumor formation through NKG2D-NKG2DL interactions
Abstract
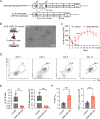
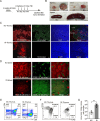
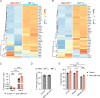
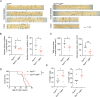
Epstein-Barr virus (EBV)-infected B cells effectively induce T cell-mediated immune surveillance that suppresses the proliferation of EBV+ B cells and development of lymphomas. However, it remains unclear whether EBV-specific T cells are involved in the surveillance of EBV-negative general tumors. To address this issue, we induced immune surveillance by expressing key EBV antigens, LMP1 and LMP2A, in germinal center B cells and investigated the formation of non-B cell tumors. LMP1/2A mice showed a significantly reduced incidence of radiation-induced T-cell acute lymphoblastic leukemia/lymphoma (T-ALL) even in the absence of LMP antigens in tumor cells and an extended life-span compared to control mice. LMP1/2A mice showed significantly higher numbers of activated memory T cells in both CD4+ and CD8+ αβT cell fractions compared to controls, suggesting their role in the elimination of tumor cells. Despite nearly absent MHC class I expression, tumor cells were effectively killed by CD8+ T cells activated upon LMP1/2A-expressing B cells. Transcriptome analysis identified upregulation of the NKG2D-NKG2DL pathway, emphasizing the capacity of LMP1/2A-induced T cells in the recognition of common tumor specific antigens. Moreover, not only T-cell tumors, but also intestinal tumors caused by ApcMin mutation were significantly suppressed by the LMP1/2A-induced immune surveillance. These results suggest that LMP1/2A-expression associated with EBV infection contributes to pan-tumor surveillance, implicating a beneficial aspect of EBV infection in humans and providing important insights into cancer prevention.
Keywords: ApcMin; Epstein-Barr virus; LMP1; LMP2A; NKG2D; T-ALL; immune surveillance; radiation-induced tumor.
Copyright © 2025 Jin, Guo, Kawano, Sasatani, Ohki, Yamane, Ota, Tamura, Sotomaru, Baba and Yasuda.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Similar articles
-
Epstein-Barr virus latent membrane protein 1 subverts IMPDH pathways to drive B-cell oncometabolism.PLoS Pathog. 2025 May 14;21(5):e1013092. doi: 10.1371/journal.ppat.1013092. eCollection 2025 May. PLoS Pathog. 2025. PMID: 40367275 Free PMC article.
-
Genetic variants in NKG2D axis and susceptibility to Epstein-Barr virus-induced nasopharyngeal carcinoma.J Cancer Res Clin Oncol. 2021 Mar;147(3):713-723. doi: 10.1007/s00432-020-03475-5. Epub 2021 Jan 3. J Cancer Res Clin Oncol. 2021. PMID: 33392659 Free PMC article.
-
Epstein-Barr virus-infected tonsillar marginal zone B cells in vivo as a precursor for immunosuppression-related B-cell lymphoma.J Virol. 2025 Aug 19;99(8):e0105124. doi: 10.1128/jvi.01051-24. Epub 2025 Jul 8. J Virol. 2025. PMID: 40626690 Free PMC article.
-
The PD-1/PD-L1 pathway and Epstein-Barr virus.Eur J Med Res. 2025 Jun 18;30(1):486. doi: 10.1186/s40001-025-02694-1. Eur J Med Res. 2025. PMID: 40533842 Free PMC article. Review.
-
HIF-1α: A Key Factor Mediating Tumor Cells from Digestive System to Evade NK Cell Killing via Activating Metalloproteinases to Hydrolyze MICA/B.Biomolecules. 2025 Jun 19;15(6):899. doi: 10.3390/biom15060899. Biomolecules. 2025. PMID: 40563539 Free PMC article. Review.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

